Project Team ID = PTID-CDS-JUL21-1171 (Members - Diana, Hema, Pavithra and Sophiya)
Project ID = PRCP-1001-RiceLeaf (Automobile Imports Price Prediction)
—— Preliminary → Identify The Business Case ——
This is an Automobile Imports Data Set which contains various characteristics of the cars including insurance risk rating and other price indicators. The target is to predict the price of the cars. Consequently, in order to avoid ‘the curse of dimensionality’, we’re gonna involve the top 10 of most influence features and will involve it to be a part of prediction journey (passing 10 selected features into X).
Because of main focus is to finding the most vigorous correlation that affecting price (target), the most opportune method to solve this case is by applying regression.
#Importing Important Libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import LabelEncoder
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LinearRegression
from sklearn.metrics import r2_score
from statsmodels.stats.outliers_influence import variance_inflation_factor
from sklearn.linear_model import Ridge
from sklearn.linear_model import Lasso
from sklearn.svm import SVR
from sklearn.model_selection import GridSearchCV
from sklearn.decomposition import PCA
import pickle
#Loading the dataset from .csv file, & rename each columns (as stated in the .txt file)
data = pd.read_csv('auto_imports.csv', names=['symboling', 'normalized_losses', 'make', 'fuel_type', 'aspiration',
'num_of_doors', 'body_style', 'drive_wheels', 'engine_location', 'wheel_base',
'length', 'width', 'height', 'curb_weight', 'engine_type', 'num_of_cylinders',
'engine_size', 'fuel_system', 'bore', 'stroke', 'compression_ratio', 'horsepower',
'peak_rpm', 'city_mpg', 'highway_mpg', 'price'])
#show the data head
data.head().T
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| symboling | 3 | 3 | 1 | 2 | 2 |
| normalized_losses | ? | ? | ? | 164 | 164 |
| make | alfa-romero | alfa-romero | alfa-romero | audi | audi |
| fuel_type | gas | gas | gas | gas | gas |
| aspiration | std | std | std | std | std |
| num_of_doors | two | two | two | four | four |
| body_style | convertible | convertible | hatchback | sedan | sedan |
| drive_wheels | rwd | rwd | rwd | fwd | 4wd |
| engine_location | front | front | front | front | front |
| wheel_base | 88.6 | 88.6 | 94.5 | 99.8 | 99.4 |
| length | 168.8 | 168.8 | 171.2 | 176.6 | 176.6 |
| width | 64.1 | 64.1 | 65.5 | 66.2 | 66.4 |
| height | 48.8 | 48.8 | 52.4 | 54.3 | 54.3 |
| curb_weight | 2548 | 2548 | 2823 | 2337 | 2824 |
| engine_type | dohc | dohc | ohcv | ohc | ohc |
| num_of_cylinders | four | four | six | four | five |
| engine_size | 130 | 130 | 152 | 109 | 136 |
| fuel_system | mpfi | mpfi | mpfi | mpfi | mpfi |
| bore | 3.47 | 3.47 | 2.68 | 3.19 | 3.19 |
| stroke | 2.68 | 2.68 | 3.47 | 3.40 | 3.40 |
| compression_ratio | 9.0 | 9.0 | 9.0 | 10.0 | 8.0 |
| horsepower | 111 | 111 | 154 | 102 | 115 |
| peak_rpm | 5000 | 5000 | 5000 | 5500 | 5500 |
| city_mpg | 21 | 21 | 19 | 24 | 18 |
| highway_mpg | 27 | 27 | 26 | 30 | 22 |
| price | 13495 | 16500 | 16500 | 13950 | 17450 |
Predicated on the results above, the independent variable a.k.a X falls on the first row to 25 (in this case start from: 'symboling' - 'highway_mpg'). The last row is the dependent variable a.k.a Y (price).
——————— Dataset Information ———————
Description: This data set consists of three types of entities:
- The specification of an auto in terms of various characteristics,
- Its assigned insurance risk rating,
- Its normalized losses in use as compared to other cars. The second rating corresponds to the degree to which the auto is more risky than its price indicates. Cars are initially assigned a risk factor symbol associated with its price. Then, if it is more risky (or less), this symbol is adjusted by moving it up (or down) the scale. Actuarians call this process “symboling”.
A value of +3 indicates that the auto is risky, -3 that it is probably pretty safe.
The third factor is the relative average loss payment per insured vehicle year. This value is normalized for all autos within a particular size classification (two-door small, station wagons, sports/speciality, etc…), and represents the average loss per car per year.
Note: Several of the attributes in the database could be used as a “class” attribute.
- symboling: -3, -2, -1, 0, 1, 2, 3.
- normalized-losses: continuous from 65 to 256.
- make: alfa-romero, audi, bmw, chevrolet, dodge, honda, isuzu, jaguar, mazda, mercedes-benz, mercury, mitsubishi, nissan, peugot, plymouth, porsche, renault, saab, subaru, toyota, volkswagen, volvo
- fuel-type: diesel, gas.
- aspiration: std, turbo.
- num-of-doors: four, two.
- body-style: hardtop, wagon, sedan, hatchback, convertible.
- drive-wheels: 4wd, fwd, rwd.
- engine-location: front, rear.
- wheel-base: continuous from 86.6 120.9.
- length: continuous from 141.1 to 208.1.
- width: continuous from 60.3 to 72.3.
- height: continuous from 47.8 to 59.8.
- curb-weight: continuous from 1488 to 4066.
- engine-type: dohc, dohcv, l, ohc, ohcf, ohcv, rotor.
- num-of-cylinders: eight, five, four, six, three, twelve, two.
- engine-size: continuous from 61 to 326.
- fuel-system: 1bbl, 2bbl, 4bbl, idi, mfi, mpfi, spdi, spfi.
- bore: continuous from 2.54 to 3.94.
- stroke: continuous from 2.07 to 4.17.
- compression-ratio: continuous from 7 to 23.
- horsepower: continuous from 48 to 288.
- peak-rpm: continuous from 4150 to 6600.
- city-mpg: continuous from 13 to 49.
- highway-mpg: continuous from 16 to 54.
- price: continuous from 5118 to 45400.
print("____________________________________________")
print(" Info of The Dataset")
print("____________________________________________")
data.info()
____________________________________________
Info of The Dataset
____________________________________________
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 201 entries, 0 to 200
Data columns (total 26 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 symboling 201 non-null int64
1 normalized_losses 201 non-null object
2 make 201 non-null object
3 fuel_type 201 non-null object
4 aspiration 201 non-null object
5 num_of_doors 201 non-null object
6 body_style 201 non-null object
7 drive_wheels 201 non-null object
8 engine_location 201 non-null object
9 wheel_base 201 non-null float64
10 length 201 non-null float64
11 width 201 non-null float64
12 height 201 non-null float64
13 curb_weight 201 non-null int64
14 engine_type 201 non-null object
15 num_of_cylinders 201 non-null object
16 engine_size 201 non-null int64
17 fuel_system 201 non-null object
18 bore 201 non-null object
19 stroke 201 non-null object
20 compression_ratio 201 non-null float64
21 horsepower 201 non-null object
22 peak_rpm 201 non-null object
23 city_mpg 201 non-null int64
24 highway_mpg 201 non-null int64
25 price 201 non-null int64
dtypes: float64(5), int64(6), object(15)
memory usage: 41.0+ KB
print("____________________________________________")
print(" Counting Missing Values of The Dataset")
print("____________________________________________")
data.isnull().sum()
____________________________________________
Counting Missing Values of The Dataset
____________________________________________
symboling 0
normalized_losses 0
make 0
fuel_type 0
aspiration 0
num_of_doors 0
body_style 0
drive_wheels 0
engine_location 0
wheel_base 0
length 0
width 0
height 0
curb_weight 0
engine_type 0
num_of_cylinders 0
engine_size 0
fuel_system 0
bore 0
stroke 0
compression_ratio 0
horsepower 0
peak_rpm 0
city_mpg 0
highway_mpg 0
price 0
dtype: int64
Remarks - Several of columns have missing values, they are not being captured in the above as they are considered as strings (‘?’) and not missing values. To rectify it, superseding '?' with the avail of np.NaN
Replace ‘?’ with np.NaN, so the missing values can be filled.
data = data.replace('?', np.NaN)
data.head()
| symboling | normalized_losses | make | fuel_type | aspiration | num_of_doors | body_style | drive_wheels | engine_location | wheel_base | ... | engine_size | fuel_system | bore | stroke | compression_ratio | horsepower | peak_rpm | city_mpg | highway_mpg | price | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 3 | NaN | alfa-romero | gas | std | two | convertible | rwd | front | 88.6 | ... | 130 | mpfi | 3.47 | 2.68 | 9.0 | 111 | 5000 | 21 | 27 | 13495 |
| 1 | 3 | NaN | alfa-romero | gas | std | two | convertible | rwd | front | 88.6 | ... | 130 | mpfi | 3.47 | 2.68 | 9.0 | 111 | 5000 | 21 | 27 | 16500 |
| 2 | 1 | NaN | alfa-romero | gas | std | two | hatchback | rwd | front | 94.5 | ... | 152 | mpfi | 2.68 | 3.47 | 9.0 | 154 | 5000 | 19 | 26 | 16500 |
| 3 | 2 | 164 | audi | gas | std | four | sedan | fwd | front | 99.8 | ... | 109 | mpfi | 3.19 | 3.40 | 10.0 | 102 | 5500 | 24 | 30 | 13950 |
| 4 | 2 | 164 | audi | gas | std | four | sedan | 4wd | front | 99.4 | ... | 136 | mpfi | 3.19 | 3.40 | 8.0 | 115 | 5500 | 18 | 22 | 17450 |
5 rows × 26 columns
Checking for the no. of missing values in each column
#displays features that have missing values in them
def missing_zero_values_table(data):
zero_val = (data == 0.00).astype(int).sum(axis=0)
mis_val = data.isnull().sum()
mis_val_percent = 100 * data.isnull().sum() / len(data)
mz_table = pd.concat([mis_val, mis_val_percent], axis=1)
mz_table = mz_table.rename(
columns = {0 : 'Sum of NaN', 1 : 'Total Values (%)'})
mz_table['Data Type'] = data.dtypes
mz_table = mz_table[
mz_table.iloc[:,1] != 0].sort_values(
'Total Values (%)', ascending=False).round(1)
print ("================= Brief Report =================\nData has " + str(data.shape[1]) + " columns and " + str(data.shape[0]) + " Rows.\n"
"There are " + str(mz_table.shape[0]) +
" columns that have missing values as\nrepresented below:")
return mz_table
missing_zero_values_table(data)
================= Brief Report =================
Data has 26 columns and 201 Rows.
There are 6 columns that have missing values as
represented below:
| Sum of NaN | Total Values (%) | Data Type | |
|---|---|---|---|
| normalized_losses | 37 | 18.4 | object |
| bore | 4 | 2.0 | object |
| stroke | 4 | 2.0 | object |
| num_of_doors | 2 | 1.0 | object |
| horsepower | 2 | 1.0 | object |
| peak_rpm | 2 | 1.0 | object |
#slice vars contains NaN values
slice_var = data[['normalized_losses', 'bore', 'stroke', 'num_of_doors', 'horsepower', 'peak_rpm']]
slice_var
#visualize it
plt.rcdefaults()
fig, ax = plt.subplots()
#columns contain missing values
var = ('normalized_losses', 'bore', 'stroke', 'num_of_doors', 'horsepower', 'peak_rpm')
y_pos = np.arange(len(var))
perc = 100 * slice_var.isnull().sum() / len(slice_var)
ax.barh(y_pos, perc, align = 'center')
ax.set_yticks(y_pos)
ax.set_yticklabels(slice_var)
ax.invert_yaxis() # labels read top-to-bottom
ax.set_xlabel('Total Values (%)', fontweight='bold')
ax.set_ylabel('Variables', fontweight='bold')
ax.set_title('Brief Report', size = 20)
plt.savefig('[Fig 2.1] Brief Report.png', dpi = 300)
plt.show()
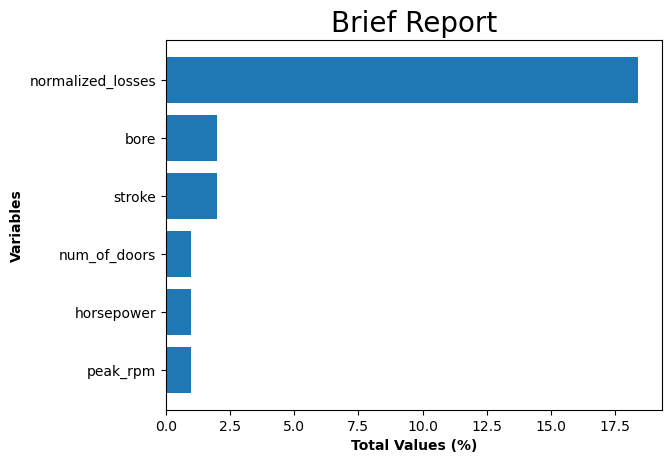
Interpretation:
- There are 6 columns that have missing values.
- Some of the columns are recorded as ‘object’ although they have values that are continuous.
- Switching ‘object’ dtype to numeric for 5 columns (excluding ‘num_of_doors’) which is categorical variable.
data[['normalized_losses', 'bore', 'stroke', 'horsepower', 'peak_rpm']] = data[['normalized_losses', 'bore', 'stroke', 'horsepower', 'peak_rpm']].apply(pd.to_numeric)
# For the numerical values, replacing the missing values with mean of their respective columns
data['normalized_losses'].fillna(data['normalized_losses'].mean(), inplace = True)
data['bore'].fillna(data['bore'].mean(), inplace = True)
data['stroke'].fillna(data['stroke'].mean(), inplace = True)
data['horsepower'].fillna(data['horsepower'].mean(), inplace = True)
data['peak_rpm'].fillna(data['peak_rpm'].mean(), inplace = True)
# For the categorical value (num_of_doors), replacing the missing values with most frequently appearing values (mode)
imputer = SimpleImputer(strategy='most_frequent')
data.iloc[:,:] = imputer.fit_transform(data)
# re-execute previous functions (missing values)
missing_zero_values_table(data)
================= Brief Report =================
Data has 26 columns and 201 Rows.
There are 0 columns that have missing values as
represented below:
| Sum of NaN | Total Values (%) | Data Type |
|---|
Missing Values has been completely handle.
data.describe().T
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| symboling | 201.0 | 0.840796 | 1.254802 | -2.00 | 0.00 | 1.00000 | 2.00 | 3.00 |
| normalized_losses | 201.0 | 122.000000 | 31.996250 | 65.00 | 101.00 | 122.00000 | 137.00 | 256.00 |
| wheel_base | 201.0 | 98.797015 | 6.066366 | 86.60 | 94.50 | 97.00000 | 102.40 | 120.90 |
| length | 201.0 | 174.200995 | 12.322175 | 141.10 | 166.80 | 173.20000 | 183.50 | 208.10 |
| width | 201.0 | 65.889055 | 2.101471 | 60.30 | 64.10 | 65.50000 | 66.60 | 72.00 |
| height | 201.0 | 53.766667 | 2.447822 | 47.80 | 52.00 | 54.10000 | 55.50 | 59.80 |
| curb_weight | 201.0 | 2555.666667 | 517.296727 | 1488.00 | 2169.00 | 2414.00000 | 2926.00 | 4066.00 |
| engine_size | 201.0 | 126.875622 | 41.546834 | 61.00 | 98.00 | 120.00000 | 141.00 | 326.00 |
| bore | 201.0 | 3.330711 | 0.268072 | 2.54 | 3.15 | 3.31000 | 3.58 | 3.94 |
| stroke | 201.0 | 3.256904 | 0.316048 | 2.07 | 3.11 | 3.29000 | 3.41 | 4.17 |
| compression_ratio | 201.0 | 10.164279 | 4.004965 | 7.00 | 8.60 | 9.00000 | 9.40 | 23.00 |
| horsepower | 201.0 | 103.396985 | 37.365602 | 48.00 | 70.00 | 95.00000 | 116.00 | 262.00 |
| peak_rpm | 201.0 | 5117.587940 | 478.113178 | 4150.00 | 4800.00 | 5117.58794 | 5500.00 | 6600.00 |
| city_mpg | 201.0 | 25.179104 | 6.423220 | 13.00 | 19.00 | 24.00000 | 30.00 | 49.00 |
| highway_mpg | 201.0 | 30.686567 | 6.815150 | 16.00 | 25.00 | 30.00000 | 34.00 | 54.00 |
| price | 201.0 | 13207.129353 | 7947.066342 | 5118.00 | 7775.00 | 10295.00000 | 16500.00 | 45400.00 |
Encoding the categorical variables for model building using LabelEncoder
label = LabelEncoder()
data1 = data.apply(label.fit_transform)
data1.head()
| symboling | normalized_losses | make | fuel_type | aspiration | num_of_doors | body_style | drive_wheels | engine_location | wheel_base | ... | engine_size | fuel_system | bore | stroke | compression_ratio | horsepower | peak_rpm | city_mpg | highway_mpg | price | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 5 | 28 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 2 | ... | 18 | 5 | 24 | 4 | 13 | 31 | 10 | 8 | 10 | 117 |
| 1 | 5 | 28 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 2 | ... | 18 | 5 | 24 | 4 | 13 | 31 | 10 | 8 | 10 | 136 |
| 2 | 3 | 28 | 0 | 1 | 0 | 1 | 2 | 2 | 0 | 10 | ... | 28 | 5 | 1 | 28 | 13 | 46 | 10 | 6 | 9 | 136 |
| 3 | 4 | 42 | 1 | 1 | 0 | 0 | 3 | 1 | 0 | 29 | ... | 11 | 5 | 13 | 25 | 22 | 27 | 17 | 11 | 13 | 122 |
| 4 | 4 | 42 | 1 | 1 | 0 | 0 | 3 | 0 | 0 | 28 | ... | 22 | 5 | 13 | 25 | 5 | 34 | 17 | 5 | 5 | 147 |
5 rows × 26 columns
Cross Checking if the label encoder has encoded all the values properly
#head of dataset (data types 'object' only)
data.select_dtypes(include = 'object').head()
| make | fuel_type | aspiration | num_of_doors | body_style | drive_wheels | engine_location | engine_type | num_of_cylinders | fuel_system | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | alfa-romero | gas | std | two | convertible | rwd | front | dohc | four | mpfi |
| 1 | alfa-romero | gas | std | two | convertible | rwd | front | dohc | four | mpfi |
| 2 | alfa-romero | gas | std | two | hatchback | rwd | front | ohcv | six | mpfi |
| 3 | audi | gas | std | four | sedan | fwd | front | ohc | four | mpfi |
| 4 | audi | gas | std | four | sedan | 4wd | front | ohc | five | mpfi |
print('-------------------------------------------------------- 1. [make] ----------------------------------------------------------')
print('Categorical Values: ',data.make.unique())
print('Numerical Values: ',data1.make.unique(), '\n')
print('----------------------------------------------------- 2. [fuel_type] --------------------------------------------------------')
print('Categorical Values: ',data.fuel_type.unique())
print('Numerical Values: ',data1.fuel_type.unique(), '\n')
print('----------------------------------------------------- 3. [aspiration] --------------------------------------------------------')
print('Categorical Values: ',data.aspiration.unique())
print('Numerical Values: ',data1.aspiration.unique(), '\n')
print('---------------------------------------------------- 4. [num_of_doors] -------------------------------------------------------')
print('Categorical Values: ',data.num_of_doors.unique())
print('Numerical Values: ',data1.num_of_doors.unique(), '\n')
print('----------------------------------------------------- 5. [body_style] --------------------------------------------------------')
print('Categorical Values: ',data.body_style.unique())
print('Numerical Values: ',data1.body_style.unique(), '\n')
print('---------------------------------------------------- 6. [drive_wheels] -------------------------------------------------------')
print('Categorical Values: ',data.drive_wheels.unique())
print('Numerical Values: ',data1.drive_wheels.unique(), '\n')
print('--------------------------------------------------- 7. [engine_location] ------------------------------------------------------')
print('Categorical Values: ',data.engine_location.unique())
print('Numerical Values: ',data1.engine_location.unique(),'\n')
print('----------------------------------------------------- 8. [engine_type] --------------------------------------------------------')
print('Categorical Values: ',data.engine_type.unique())
print('Numerical Values: ',data1.engine_type.unique(),'\n')
print('-------------------------------------------------- 9. [num_of_cylinders] -----------------------------------------------------')
print('Categorical Values: ',data.num_of_cylinders.unique())
print('Numerical Values: ',data1.num_of_cylinders.unique(),'\n')
print('---------------------------------------------------- 10. [fuel_system] -------------------------------------------------------')
print('Categorical Values: ',data.fuel_system.unique())
print('Numerical Values: ',data1.fuel_system.unique(), '\n')
-------------------------------------------------------- 1. [make] ----------------------------------------------------------
Categorical Values: ['alfa-romero' 'audi' 'bmw' 'chevrolet' 'dodge' 'honda' 'isuzu' 'jaguar'
'mazda' 'mercedes-benz' 'mercury' 'mitsubishi' 'nissan' 'peugot'
'plymouth' 'porsche' 'renault' 'saab' 'subaru' 'toyota' 'volkswagen'
'volvo']
Numerical Values: [ 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21]
----------------------------------------------------- 2. [fuel_type] --------------------------------------------------------
Categorical Values: ['gas' 'diesel']
Numerical Values: [1 0]
----------------------------------------------------- 3. [aspiration] --------------------------------------------------------
Categorical Values: ['std' 'turbo']
Numerical Values: [0 1]
---------------------------------------------------- 4. [num_of_doors] -------------------------------------------------------
Categorical Values: ['two' 'four']
Numerical Values: [1 0]
----------------------------------------------------- 5. [body_style] --------------------------------------------------------
Categorical Values: ['convertible' 'hatchback' 'sedan' 'wagon' 'hardtop']
Numerical Values: [0 2 3 4 1]
---------------------------------------------------- 6. [drive_wheels] -------------------------------------------------------
Categorical Values: ['rwd' 'fwd' '4wd']
Numerical Values: [2 1 0]
--------------------------------------------------- 7. [engine_location] ------------------------------------------------------
Categorical Values: ['front' 'rear']
Numerical Values: [0 1]
----------------------------------------------------- 8. [engine_type] --------------------------------------------------------
Categorical Values: ['dohc' 'ohcv' 'ohc' 'l' 'rotor' 'ohcf']
Numerical Values: [0 4 2 1 5 3]
-------------------------------------------------- 9. [num_of_cylinders] -----------------------------------------------------
Categorical Values: ['four' 'six' 'five' 'three' 'twelve' 'two' 'eight']
Numerical Values: [2 3 1 4 5 6 0]
---------------------------------------------------- 10. [fuel_system] -------------------------------------------------------
Categorical Values: ['mpfi' '2bbl' 'mfi' '1bbl' 'spfi' '4bbl' 'idi' 'spdi']
Numerical Values: [5 1 4 0 7 2 3 6]
# splitting the data into X and Y so we can do feature selection
x = data1.drop('price', axis=1)
y = data1['price']
print(x.head())
y.head()
symboling normalized_losses make fuel_type aspiration num_of_doors \
0 5 28 0 1 0 1
1 5 28 0 1 0 1
2 3 28 0 1 0 1
3 4 42 1 1 0 0
4 4 42 1 1 0 0
body_style drive_wheels engine_location wheel_base ... \
0 0 2 0 2 ...
1 0 2 0 2 ...
2 2 2 0 10 ...
3 3 1 0 29 ...
4 3 0 0 28 ...
num_of_cylinders engine_size fuel_system bore stroke \
0 2 18 5 24 4
1 2 18 5 24 4
2 3 28 5 1 28
3 2 11 5 13 25
4 1 22 5 13 25
compression_ratio horsepower peak_rpm city_mpg highway_mpg
0 13 31 10 8 10
1 13 31 10 8 10
2 13 46 10 6 9
3 22 27 17 11 13
4 5 34 17 5 5
[5 rows x 25 columns]
0 117
1 136
2 136
3 122
4 147
Name: price, dtype: int64
Remarks - Data is now cleaned (missing values are filled and all the categorical variables are, and ready for analysis.)
Disclaimer - One hot encoder etc. were not used although there is a risk of the encoded values being ranked based on the encoded values and may be considered to be more important than the others in the regression analysis.
#saving cleaned data to .csv files
data1.to_csv('auto_imports [cleaned].csv', index=False)
At this time we have chosen not to add more variables (over 50) into the mix with the other forms of encoding as we have performed feature selection analysis to reduce the dimensions for modeling. In the event any of the categorical variables are selected as an important feature by the feature selection technique then we would use other forms of encoding and add the new encoded variables to the predictor data set to do the modeling.
# FEATURE SELECTION TECHNIQUE
#apply SelectKBest class to extract top 10 best features
bestfeatures = SelectKBest(score_func=chi2, k=10)
fit = bestfeatures.fit(x,y)
dfscores = pd.DataFrame(fit.scores_)
dfcolumns = pd.DataFrame(x.columns)
#concat two dataframes for better visualization
featurescores = pd.concat([dfcolumns,dfscores], axis=1)
featurescores.columns = ['Specs', 'Score'] #naming the data
print(featurescores.nlargest(10, 'Score'))
Specs Score
13 curb_weight 5691.994803
10 length 2319.534038
21 horsepower 1865.324072
9 wheel_base 1657.488294
12 height 1435.530316
16 engine_size 1367.586605
1 normalized_losses 1285.230854
11 width 1277.660011
18 bore 1124.427128
19 stroke 790.046992
The top 10 features are now used to build the models for comparison
Modifying The Dataset by Including These 10 Selected Features
X = x[['curb_weight', 'length', 'horsepower','wheel_base','height','engine_size','normalized_losses','width','bore','stroke']].copy()
X.head()
| curb_weight | length | horsepower | wheel_base | height | engine_size | normalized_losses | width | bore | stroke | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 92 | 22 | 31 | 2 | 1 | 18 | 28 | 8 | 24 | 4 |
| 1 | 92 | 22 | 31 | 2 | 1 | 18 | 28 | 8 | 24 | 4 |
| 2 | 115 | 29 | 46 | 10 | 13 | 28 | 28 | 16 | 1 | 28 |
| 3 | 66 | 44 | 27 | 29 | 25 | 11 | 42 | 21 | 13 | 25 |
| 4 | 116 | 44 | 34 | 28 | 25 | 22 | 42 | 23 | 13 | 25 |
# Checking for the skewness and kurtosis for 10 selected features and target
print("Skewness of curb_weight: %f" %X['curb_weight'].skew())
print("Kurtosis of curb_weight: %f" %X['curb_weight'].kurt())
print("Skewness of length: %f" %X['length'].skew())
print("Kurtosis of length: %f" %X['length'].kurt())
print("Skewness of horsepower: %f" %X['horsepower'].skew())
print("Kurtosis of horsepower: %f" %X['horsepower'].kurt())
print("Skewness of wheel_base: %f" %X['wheel_base'].skew())
print("Kurtosis of wheel_base: %f" %X['wheel_base'].kurt())
print("Skewness of height: %f" %X['height'].skew())
print("Kurtosis of height: %f" %X['height'].kurt())
print("Skewness of engine_size: %f" %X['engine_size'].skew())
print("Kurtosis of engine_size %f" %X['engine_size'].kurt())
print("Skewness of normalized_losses: %f" %X['normalized_losses'].skew())
print("Kurtosis of normalized_losses: %f" %X['normalized_losses'].kurt())
print("Skewness of width: %f" %X['width'].skew())
print("Kurtosis of width: %f" %X['width'].kurt())
print("Skewness of bore: %f" %X['bore'].skew())
print("Kurtosis of bore: %f" %X['bore'].kurt())
print("Skewness of stroke: %f" %X['stroke'].skew())
print("Kurtosis of stroke: %f" %X['stroke'].kurt())
print("Skewness of Price: %f" %data1['price'].skew())
print("Kurtosis of Price: %f" %data1['price'].kurt())
Skewness of curb_weight: 0.093721
Kurtosis of curb_weight: -1.147269
Skewness of length: 0.019671
Kurtosis of length: -1.176374
Skewness of horsepower: 0.430146
Kurtosis of horsepower: -0.886195
Skewness of wheel_base: 0.449951
Kurtosis of wheel_base: -0.955813
Skewness of height: 0.097420
Kurtosis of height: -1.143859
Skewness of engine_size: 0.623072
Kurtosis of engine_size -0.766610
Skewness of normalized_losses: -0.176570
Kurtosis of normalized_losses: -0.842252
Skewness of width: 0.381469
Kurtosis of width: -0.888932
Skewness of bore: 0.131851
Kurtosis of bore: -1.187268
Skewness of stroke: -0.368742
Kurtosis of stroke: -0.759355
Skewness of Price: 0.066486
Kurtosis of Price: -1.204690
Reference: Skewness between -0.5 and 0.5: data fairly symmetrical. Skewness between -1 and – 0.5 or between 0.5 and 1: data moderately skewed. Skewness < -1 or > than 1: the data are highly skewed. Kurtosis between -2 and +2 are considered acceptable.
Teams Remarks: The Skewness and Kurtosis for all the predictor and target variables falls within the acceptable range.
#Boxplot analysis
figure, ax = plt.subplots(2,5, figsize=(20,10))
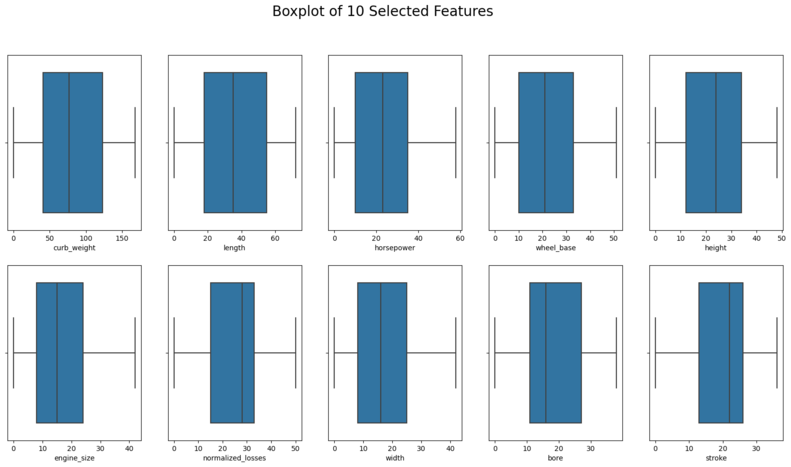
plt.suptitle('Boxplot of 10 Selected Features', size = 20)
sns.boxplot(data1['curb_weight'],ax=ax[0,0])
sns.boxplot(data1['length'], ax=ax[0,1])
sns.boxplot(data1['horsepower'], ax=ax[0,2])
sns.boxplot(data1['wheel_base'], ax=ax[0,3])
sns.boxplot(data1['height'], ax=ax[0,4])
sns.boxplot(data1['engine_size'], ax=ax[1,0])
sns.boxplot(data1['normalized_losses'], ax=ax[1,1])
sns.boxplot(data1['width'], ax=ax[1,2])
sns.boxplot(data1['bore'], ax=ax[1,3])
sns.boxplot(data1['stroke'], ax=ax[1,4])
plt.savefig('[Fig 3.1] Boxplot of 10 Selected Features.png', dpi = 300)
plt.show()
figure, ax = plt.subplots(2,5, figsize=(20,10))
#See the distribution of the data
plt.suptitle('Distribution of 10 Selected Features', size = 20)
sns.distplot(data1['curb_weight'],ax=ax[0,0])
sns.distplot(data1['length'], ax=ax[0,1])
sns.distplot(data1['horsepower'], ax=ax[0,2])
sns.distplot(data1['wheel_base'], ax=ax[0,3])
sns.distplot(data1['height'], ax=ax[0,4])
sns.distplot(data1['engine_size'], ax=ax[1,0])
sns.distplot(data1['normalized_losses'], ax=ax[1,1])
sns.distplot(data1['width'], ax=ax[1,2])
sns.distplot(data1['bore'], ax=ax[1,3])
sns.distplot(data1['stroke'], ax=ax[1,4])
plt.savefig('[Fig 3.2] Distribution of 10 Selected Features.png', dpi = 300)
plt.show()
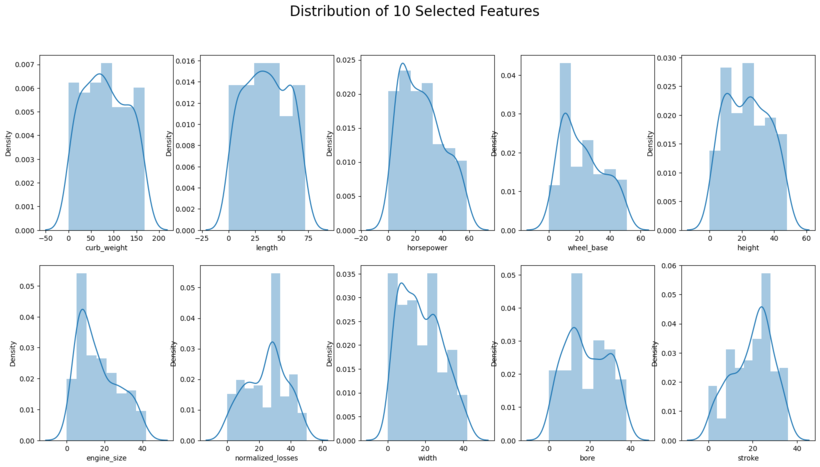
Interpretation: Boxplot and distribution plots look fine.
#combining the x (features) and y (target) to show the heatmap and pairplot analysis
xydata = data1[['curb_weight', 'length', 'horsepower','wheel_base','height','engine_size','normalized_losses','width','bore','stroke', 'price']].copy()
xydata.head()
| curb_weight | length | horsepower | wheel_base | height | engine_size | normalized_losses | width | bore | stroke | price | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 92 | 22 | 31 | 2 | 1 | 18 | 28 | 8 | 24 | 4 | 117 |
| 1 | 92 | 22 | 31 | 2 | 1 | 18 | 28 | 8 | 24 | 4 | 136 |
| 2 | 115 | 29 | 46 | 10 | 13 | 28 | 28 | 16 | 1 | 28 | 136 |
| 3 | 66 | 44 | 27 | 29 | 25 | 11 | 42 | 21 | 13 | 25 | 122 |
| 4 | 116 | 44 | 34 | 28 | 25 | 22 | 42 | 23 | 13 | 25 | 147 |
#Heatmap to shows the correlation
plt.figure(figsize=(20,15))
sns.heatmap(xydata.corr(),cmap='nipy_spectral',annot=True)
plt.title('Heatmap of 10 Selected Features and Target',
fontsize=25)
plt.savefig('[Fig 3.3] Heatmap of 10 Selected Features and Target.png', dpi = 300)
plt.show()
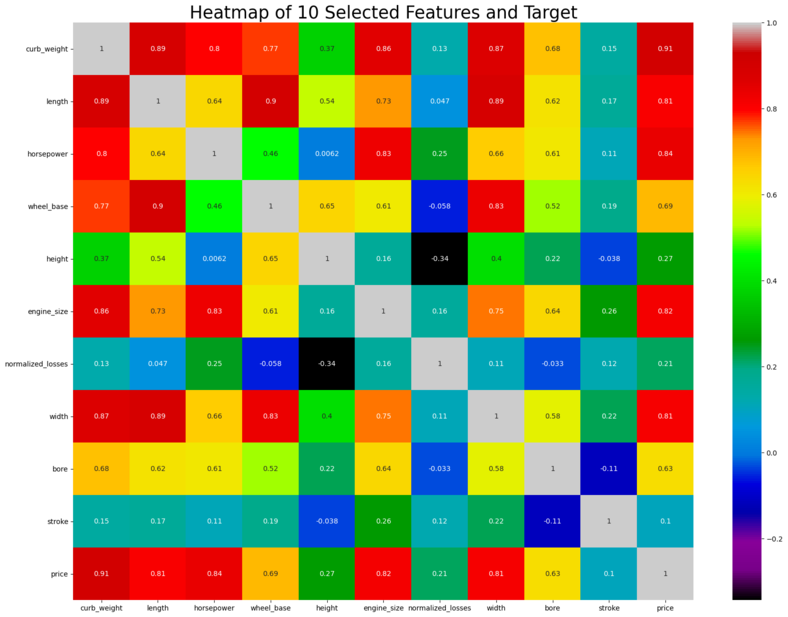
Teams Remarks: There seems to be significant amount of positive correlation between some of the predictor variables. Multicollinearity occurs when independent variables in a regression model are correlated. This correlation is a problem because independent variables should be independent. If the degree of correlation between variables is high enough, it can cause problems when we fit the model and interpret the results.
curb_weight is correlated to length, horsepower, wheelbase, enginesize, width and bore. Length is correlated to wheelbase and width. horsepower is correlated to engine size. Wheelbase is correlated to horsepower and width
Multicollinearity analysis is to be done in detail. See below
# Pairplot analysis
sns.pairplot(xydata)
plt.savefig('[Fig 3.4] Pairplot of 10 Selected Features and Target.png', dpi = 300)
plt.show()
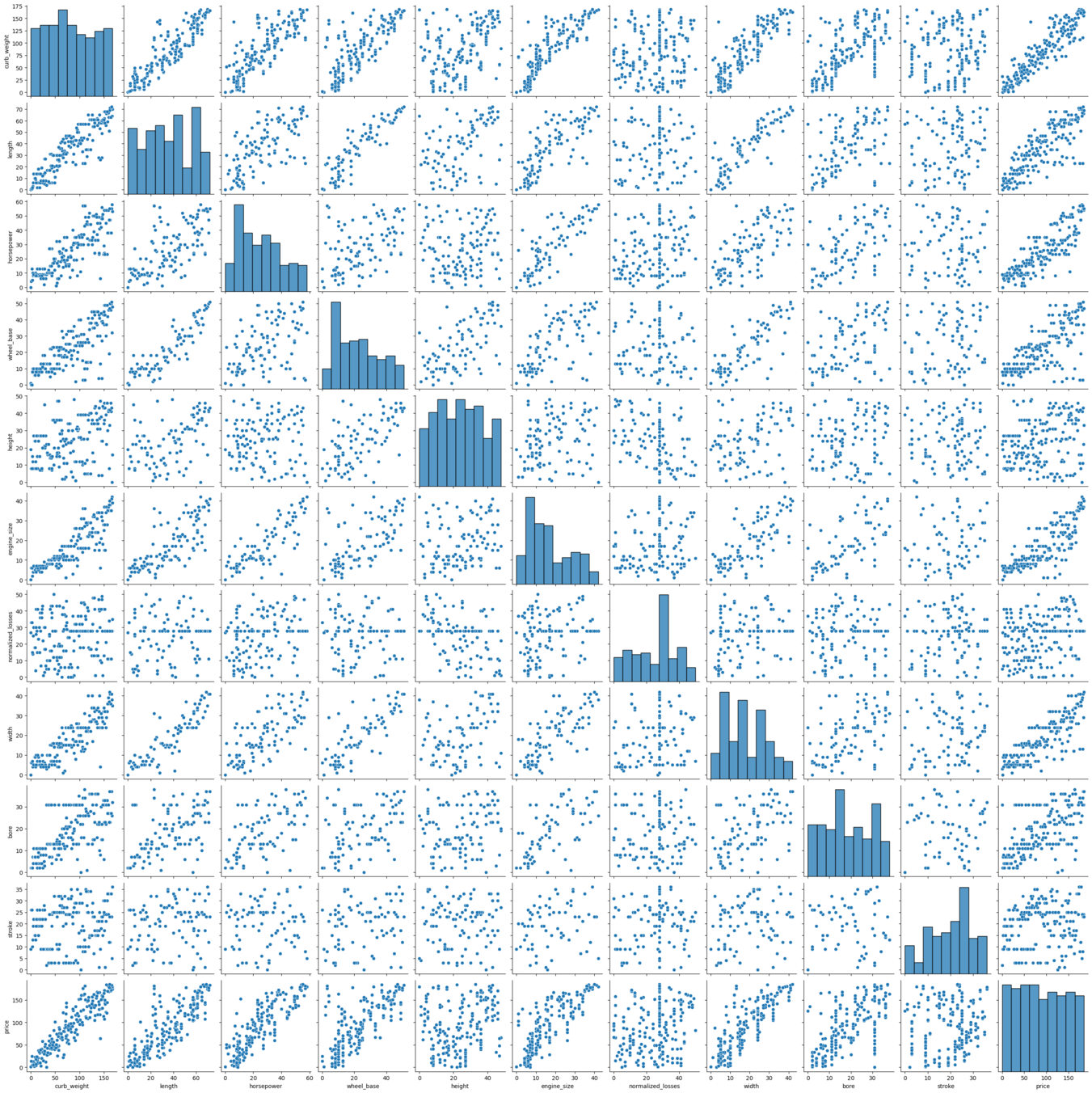
Remarks: As is evident from the pair plots, the target variable is showing a nice linear fit with some of the predictor variables. So, linear regression would provide felicitous model to the dataset.
Also there seems to be a linear relationship between some of the predictor variables, which is natural as the curb weight is dependent on the length, width and engine_size of the cars. This multicollinearity problem will be resolved by PCA analysis.
Residual plots to see if there are any patterns that could cause errors in the regression models.
figure, ax = plt.subplots(2,5, figsize=(20,10))
#See the Residual plots of the data
plt.suptitle('Residual Plot of 10 Selected Features Against Target', size = 20)
sns.residplot(data1['curb_weight'],data1['price'],ax=ax[0,0])
sns.residplot(data1['length'], data1['price'], ax=ax[0,1])
sns.residplot(data1['horsepower'], data1['price'], ax=ax[0,2])
sns.residplot(data1['wheel_base'], data1['price'], ax=ax[0,3])
sns.residplot(data1['height'], data1['price'], ax=ax[0,4])
sns.residplot(data1['engine_size'], data1['price'], ax=ax[1,0])
sns.residplot(data1['normalized_losses'], data1['price'], ax=ax[1,1])
sns.residplot(data1['width'], data1['price'], ax=ax[1,2])
sns.residplot(data1['bore'], data1['price'], ax=ax[1,3])
sns.residplot(data1['stroke'], data1['price'], ax=ax[1,4])
plt.savefig('[Fig 3.5] Residual Plot of 10 Selected Features Against Target.png', dpi = 300)
plt.show()
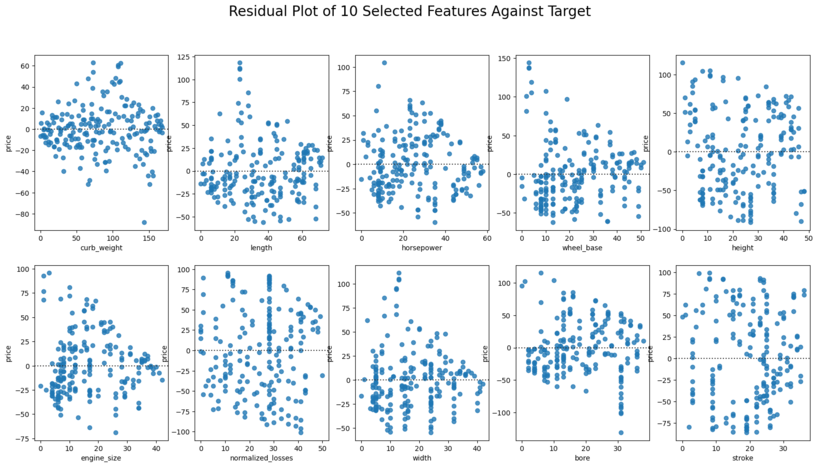
Remarks: No significant patterns in the residual plots
We vigorously believe when it comes to ML algorithm, "One Size Does Not Fit All". The same things additionally occur when it comes to modeling. To resolve the issue, several regression models will be utilized for the comparative analysis and the best model (shown by r2 score) will be chosen for this project.
Model 1: Multiple Linear Regression
Model 2: Ridge Regression
Model 3: Lasso Regression
Model 4: Support Vector Regression
Model 5: Principal Component Analysis (PCA)
Splitting the dataset into train and test data sets for validation
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)
X_train.shape, X_test.shape, y_train.shape, y_test.shape
((134, 10), (67, 10), (134,), (67,))
#feature scaling
sc = StandardScaler()
X_train_fc = sc.fit_transform(X_train)
X_test_fc = sc.transform(X_test)
Model 1: Multiple Linear Regression
# building the linear regression model
lm=LinearRegression()
# fitting the model with the standardized data
lm.fit(X_train_fc,y_train)
LinearRegression()
# predicting with the linear model
y_pred=lm.predict(X_test_fc)
Linear_Regr_score = r2_score(y_test,y_pred)
Linear_Regr_score
0.8675330801508967
Pickling The Model (Save and Load The Model)
with open('[4.1.1] Multiple_LR', 'wb') as f:
pickle.dump(lm, f)
with open('[4.1.1] Multiple_LR', 'rb') as f:
model_mlr = pickle.load(f)
#testing the pickled model. reason: to make sure the pickle output showing the same as the current model (y_pred)
model_mlr.predict(X_test_fc)
array([ 39.27873438, 172.39008554, 23.79690875, 38.81598645,
118.02560264, 24.27532209, 166.90033166, 78.36283435,
163.91513232, 182.72922042, 149.71410486, 56.22400104,
108.4322148 , 77.68539223, 113.30317396, 49.2278126 ,
82.63551223, 35.51762066, 74.26413099, 178.80950224,
125.98704081, 179.6828716 , 19.49939044, 30.30081594,
154.13705213, 74.45561159, 98.72363617, 159.9344475 ,
128.14370608, 148.09795606, 102.11328533, 17.0646285 ,
115.37359601, 38.42050953, 40.86989298, 138.82835563,
28.40460768, 53.41603648, 106.81471585, 65.3142471 ,
126.20111039, 140.32820502, 62.90172307, 34.66597309,
128.76048409, 131.86746884, 12.84386201, 17.43809555,
81.15455802, 23.63177825, 163.92478736, 162.83679969,
105.50518899, 103.04129662, 34.36741024, 31.95051374,
69.67767341, 150.74724269, 81.05866593, 74.96955417,
74.50898045, 73.35205522, 12.8711576 , 127.07701526,
28.46784132, 49.29817804, 60.93400171])
#Plot of Actual Values against Predicted Values
plt.figure(figsize=(10,5))
plt.scatter(y_test,y_pred)
plt.xlabel("actual")
plt.ylabel("predicted")
plt.title('Plot of Actual Values against Predicted Values',size = 15)
plt.savefig('[Fig 4.1] MultiLinReg Actual Against Target.png', dpi = 300)
plt.show()
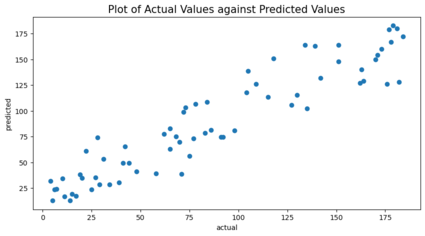
Checking to see if the score can be improved if multicollinearity issue is resolved.
The potential solutions for resolving multicollinearity issues among the predictor variables are:
- Remove some of the highly correlated independent variables.
- Linearly combine the independent variables, such as adding them together.
- Perform an analysis designed for highly correlated variables, such as principal components analysis or partial least squares regression.
- Lasso and Ridge regression are advanced forms of regression analysis that can handle multicollinearity.
Improving the model - Iteration 1
N.B: Using the first solution from the above and reducing some of the highly correlated independent variables.
Part 1 - According to heatmap visualization, removing those predictor variables that show low correlation with target variables
(stroke 0.1, normalized losses 0.21 and height 0.27), new X data set was made.
X_new = x[['curb_weight', 'length', 'horsepower','wheel_base','engine_size','width','bore']].copy()
X_new.head()
| curb_weight | length | horsepower | wheel_base | engine_size | width | bore | |
|---|---|---|---|---|---|---|---|
| 0 | 92 | 22 | 31 | 2 | 18 | 8 | 24 |
| 1 | 92 | 22 | 31 | 2 | 18 | 8 | 24 |
| 2 | 115 | 29 | 46 | 10 | 28 | 16 | 1 |
| 3 | 66 | 44 | 27 | 29 | 11 | 21 | 13 |
| 4 | 116 | 44 | 34 | 28 | 22 | 23 | 13 |
#creating train and test data set
X_trn_new, X_tst_new, y_trn_new, y_tst_new = train_test_split(X_new, y, test_size=0.33, random_state=42)
X_trn_new.shape, X_tst_new.shape, y_trn_new.shape, y_tst_new.shape
# scaling the features
X_trn_new_fc = sc.fit_transform(X_trn_new)
X_tst_new_fc = sc.transform(X_tst_new)
#fitting the model
lm.fit(X_trn_new_fc,y_trn_new)
# predicting using the test dataset
y_pred_new = lm.predict(X_tst_new_fc)
#checking the r2score using the new dataset
r2_score(y_tst_new,y_pred_new)
0.8780153528258652
Pickling The Model (Save and Load The Model)
with open('[4.1.2] Multiple_LR (Reduce Features According to Heatmap)', 'wb') as f:
pickle.dump(lm, f)
with open('[4.1.2] Multiple_LR (Reduce Features According to Heatmap)', 'rb') as f:
model_mlr = pickle.load(f)
#testing the pickled model. reason: to make sure the pickle output showing the same as the current model (y_pred_new)
model_mlr.predict(X_tst_new_fc)
array([ 29.86454227, 171.6891113 , 22.1486513 , 42.82401536,
112.11902126, 22.46252551, 167.60650606, 89.02408535,
155.05415392, 182.05624926, 158.03624213, 62.34155084,
104.18416228, 70.59730038, 113.5235974 , 47.40173449,
79.14340238, 39.43691677, 71.86982329, 177.99817272,
126.81306507, 180.64632423, 27.08085981, 27.93783444,
162.6240674 , 72.43507378, 98.88925128, 168.07217211,
129.0711308 , 144.28285008, 87.10858806, 19.29036592,
113.36858606, 31.34724299, 48.3701066 , 131.16942701,
32.76502916, 51.32203631, 102.49061298, 60.10549721,
129.04760861, 143.19642466, 71.99640675, 28.67220626,
124.26122654, 134.65816999, 10.35967481, 23.94054364,
84.37075626, 20.0346044 , 156.667191 , 153.92512106,
104.10954835, 105.78034077, 26.97865696, 24.57304582,
69.29689364, 150.09972471, 91.84666751, 77.43007886,
84.37297962, 75.73652957, 15.69246978, 115.5654485 ,
39.05066241, 41.80314572, 57.37789719])
Remarks: Score seemed to have improved slightly but no significant improvement
Improving the model - Iteration 2
N.B: only including the first four predictor variables that have the highest VIF values
VIF is used as a measure to check the strength of multicollinearity.
# VIF dataframe
vif_data = pd.DataFrame()
vif_data["feature"] = X_new.columns
# calculating VIF for each feature
vif_data["VIF"] = [variance_inflation_factor(X_new.values, i)
for i in range(len(X_new.columns))]
print(vif_data)
feature VIF
0 curb_weight 40.060008
1 length 46.569846
2 horsepower 14.498971
3 wheel_base 23.567041
4 engine_size 17.044365
5 width 22.841830
6 bore 7.273853
References: VIF’s start at 1 and have no upper limit. A value of 1 indicates that there is no correlation between this independent variable and any others. VIF’s between 1 and 5 suggest that there is a moderate correlation, but it is not severe enough to warrant corrective measures. VIF’s > 5 represent critical levels of multicollinearity where the coefficients are poorly estimated, and the p-values are questionable.
Interpretation: based on the above length seems to have the highest VIF as it is heavily correlated to the curb_weight (which makes sense).
Decision: eliminating the features, creating a new data set, re-executing the model, checking the r2score below.
# creating a leaner data set
X_lean = x[['curb_weight', 'horsepower','wheel_base', 'engine_size', 'width', 'bore']].copy()
# training and splitting the data set
X_trn_lean, X_tst_lean, y_trn_lean, y_tst_lean = train_test_split(X_lean, y, test_size=0.33, random_state=42)
print(X_trn_lean.shape, X_tst_lean.shape, y_trn_lean.shape, y_tst_lean.shape)
# scaling the data set
X_trn_lean_fc = sc.fit_transform(X_trn_lean)
X_tst_lean_fc = sc.transform(X_tst_lean)
#fitting the model
lm.fit(X_trn_lean_fc,y_trn_lean)
# predicting the test data
y_pred_lean = lm.predict(X_tst_lean_fc)
# checking the R2 score
linear_FinalRegr_score = r2_score(y_tst_lean,y_pred_lean)
linear_FinalRegr_score
(134, 6) (67, 6) (134,) (67,)
0.8798334755333546
r2score has improved slightly
Pickling The Model (Save and Load The Model)
with open('[4.1.3] Multiple_LR (Applied VIF)', 'wb') as f:
pickle.dump(lm, f)
with open('[4.1.3] Multiple_LR (Applied VIF)', 'rb') as f:
model_mlr = pickle.load(f)
#testing the pickled model. reason: to make sure the pickle output showing the same as the current model (y_pred_lean)
model_mlr.predict(X_tst_lean_fc)
array([ 30.27343398, 170.47430895, 22.73895155, 43.36469706,
109.23075799, 23.32399729, 167.69098889, 89.50820236,
154.98421213, 182.45706477, 159.56262655, 62.73896071,
103.07445564, 69.30465751, 112.65093414, 47.39745066,
77.26159754, 39.80364378, 71.08518415, 178.75513972,
126.99124146, 181.54374011, 28.54858269, 27.66585924,
164.05783674, 70.67878124, 98.51475024, 168.88227382,
129.36527698, 142.05415334, 85.75650386, 20.82564877,
114.89414291, 32.96710779, 46.94735796, 132.15850514,
34.18564324, 50.85908724, 101.293929 , 61.53487905,
129.33993144, 143.76753303, 69.19369441, 29.85259497,
127.24740036, 135.32206917, 10.81455533, 24.38670205,
83.77232445, 19.35673492, 158.22064528, 153.79719437,
103.25138019, 102.30832381, 28.07206833, 25.84500123,
69.61783216, 149.66836788, 92.47574675, 76.36617191,
85.08796628, 74.58564527, 17.33976336, 114.83458516,
42.15072667, 40.76474674, 54.94813759])
Remarks: We chose to stop the iterations here for the multiple linear regression model and considered this as the final model, as further eliminating the feature variables was reducing the R2score, as there seems to be an interaction effect that was explaining the variance in the target which we may be eliminating when we remove the features further. Hoping the other models could fix this problem
Model 2: Ridge Regression
Iteration 1
N.B.: Based on 7 predictors (curb_weight, length, horsepower, wheel_base, engine_size, width, bore)
#build the model
ridgereg = Ridge(alpha=0.001,normalize=True)
# fit the model with the train data
ridgereg.fit(X_trn_new_fc,y_trn_new)
# predict using test data
y_pred_ridge = ridgereg.predict(X_tst_new_fc)
# checking the r2score
Ridge_Regr_Score = r2_score(y_tst_new,y_pred_ridge)
Ridge_Regr_Score
0.8779583411163029
Pickling The Model (Save and Load The Model)
with open('[4.2.1] Ridge_Regression (Based on 7 Predictors)', 'wb') as f:
pickle.dump(ridgereg, f)
with open('[4.2.1] Ridge_Regression (Based on 7 Predictors)', 'rb') as f:
model_ridgereg = pickle.load(f)
#testing the pickled model. reason: to make sure the pickle output showing the same as the current model (y_pred_ridge)
model_ridgereg.predict(X_tst_new_fc)
array([ 29.90752489, 171.72841746, 22.20181554, 42.84708827,
112.16703655, 22.48890924, 167.58484068, 89.09038447,
155.06730399, 182.08807184, 158.04213467, 62.31051775,
104.28458407, 70.68581938, 113.57331795, 47.42832698,
79.27589636, 39.48390813, 71.93773756, 178.04361732,
126.84478101, 180.68248153, 27.08531586, 28.00594695,
162.62117749, 72.55338732, 98.83267793, 168.10438421,
129.08690111, 144.23367972, 87.12475397, 19.33164051,
113.14493387, 31.35336474, 48.32879357, 131.08277727,
32.80631043, 51.45232694, 102.602994 , 60.02862866,
129.03508323, 143.29940632, 72.06004431, 28.6547395 ,
124.24303215, 134.71369311, 10.43453629, 23.98383631,
84.35030435, 20.15852662, 156.56886533, 153.94624394,
104.18103799, 105.85956561, 26.97314943, 24.62700446,
69.30015797, 150.09920943, 91.89303458, 77.49413747,
84.3707146 , 75.8125474 , 15.71690855, 115.61714962,
39.00987341, 41.79998721, 57.38176397])
Iteration 2
N.B: Based on 6 predictors (curb_weight, horsepower, wheel_base, engine_size, width, bore)
#build the model
ridgereg = Ridge(alpha=0.001,normalize=True)
# fit the model with the train data
ridgereg.fit(X_trn_lean_fc,y_trn_lean)
# predict using test data
y_pred_ridge = ridgereg.predict(X_tst_lean_fc)
# checking the r2score
Ridge_RegrLn_Score = r2_score(y_tst_lean,y_pred_ridge)
Ridge_RegrLn_Score
0.8798256415283978
Pickling The Model (Save and Load The Model)
with open('[4.2.2] Ridge_Regression (Based on 6 Predictors)', 'wb') as f:
pickle.dump(ridgereg, f)
with open('[4.2.2] Ridge_Regression (Based on 6 Predictors)', 'rb') as f:
model_ridgereg = pickle.load(f)
#testing the pickled model. reason: to make sure the pickle output showing the same as the current model (y_pred_ridge)
model_ridgereg.predict(X_tst_lean_fc)
array([ 30.32662131, 170.48928952, 22.8094774 , 43.40048649,
109.21925918, 23.36990063, 167.67713925, 89.58443993,
154.99560231, 182.49630602, 159.60028807, 62.71330722,
103.15561604, 69.36999051, 112.68303047, 47.42270744,
77.35870432, 39.86040544, 71.14003103, 178.8157088 ,
127.03602951, 181.59888536, 28.58345025, 27.73084736,
164.08324879, 70.76394653, 98.44895951, 168.93017324,
129.39608354, 141.95640077, 85.74060671, 20.90053101,
114.6975337 , 33.00915999, 46.87386896, 132.08595346,
34.26054978, 50.98317342, 101.38557552, 61.4878803 ,
129.33833483, 143.88358492, 69.19994765, 29.8600492 ,
127.29982484, 135.39052009, 10.90459806, 24.44061205,
83.73845849, 19.47065827, 158.14855143, 153.8155753 ,
103.30593697, 102.32007364, 28.09000868, 25.9289979 ,
69.62788572, 149.65954822, 92.53450746, 76.40905069,
85.09779154, 74.63901017, 17.40026598, 114.87053641,
42.17201272, 40.74046487, 54.90188622])
Remarks: The r2scores are in line with the optimal linear regression model
Model 3: Lasso Regression
Iteration 1
N.B: Based on 7 predictors curb_weight, length, horsepower, wheel_base, engine_size, width, bore
#build the model
lassoreg = Lasso(alpha=0.001,normalize=True, max_iter=1e5)
# fit the model with the train data
lassoreg.fit(X_trn_new_fc,y_trn_new)
# predict using test data
y_pred_lasso = lassoreg.predict(X_tst_new_fc)
# checking the r2score
Lasso_Regr_Score = r2_score(y_tst_new,y_pred_lasso)
Lasso_Regr_Score
0.8780685237672865
Pickling The Model (Save and Load The Model)
with open('[4.3.1] Lasso_Reg (Based on 7 Predictors)', 'wb') as f:
pickle.dump(lassoreg, f)
with open('[4.3.1] Lasso_Reg (Based on 7 Predictors)', 'rb') as f:
model_lassoreg = pickle.load(f)
#testing the pickled model. reason: to make sure the pickle output showing the same as the current model (y_pred_lasso)
model_lassoreg.predict(X_tst_new_fc)
array([ 29.89909691, 171.7122689 , 22.17225647, 42.84909075,
112.10774173, 22.47060234, 167.65146263, 89.03125319,
155.02846092, 182.07067307, 158.06932612, 62.30548029,
104.26376805, 70.66230701, 113.51839097, 47.40501501,
79.20523781, 39.46954105, 71.92984543, 178.00124591,
126.91932597, 180.64709364, 27.10072847, 27.9802479 ,
162.64555789, 72.51964443, 98.81433155, 168.08907243,
129.1723591 , 144.183453 , 87.07236518, 19.30377477,
113.19615154, 31.35705911, 48.36186312, 131.08984315,
32.86033361, 51.38951228, 102.5739932 , 60.05829999,
129.09028949, 143.24085041, 72.00112987, 28.66644346,
124.32518492, 134.67160442, 10.41733855, 23.95003376,
84.36638304, 20.09463194, 156.57988916, 153.90194435,
104.11874362, 105.82057242, 26.97666861, 24.59795972,
69.32572504, 150.12381755, 91.8475446 , 77.44578553,
84.35368131, 75.75601068, 15.69421768, 115.61300113,
39.01772454, 41.81591627, 57.42549943])
Iteration 2
N.B: Based on 6 predictor variables curb_weight, horsepower, wheel_base, engine_size, width, bore
#build the model
lassoreg = Lasso(alpha=0.001,normalize=True, max_iter=1e5)
# fit the model with the train data
lassoreg.fit(X_trn_lean_fc,y_trn_lean)
# predict using test data
y_pred_lasso = lassoreg.predict(X_tst_lean_fc)
# checking the r2score
Lasso_RegrLn_Score = r2_score(y_tst_lean,y_pred_lasso)
Lasso_RegrLn_Score
0.879897132703048
Pickling The Model (Save and Load The Model)
with open('[4.3.2] Lasso_Reg (Based on 6 Predictors)', 'wb') as f:
pickle.dump(lassoreg, f)
with open('[4.3.2] Lasso_Reg (Based on 6 Predictors)', 'rb') as f:
model_lassoreg = pickle.load(f)
#testing the pickled model. reason: to make sure the pickle output showing the same as the current model (y_pred_lasso)
model_lassoreg.predict(X_tst_lean_fc)
array([ 30.30543908, 170.49202804, 22.75724277, 43.38870487,
109.21186203, 23.33063793, 167.73464662, 89.50906937,
154.95522959, 182.46814219, 159.59546561, 62.7059535 ,
103.14461365, 69.36056567, 112.6386984 , 47.3982503 ,
77.30882506, 39.83274729, 71.13854447, 178.75243779,
127.09461674, 181.54017866, 28.57069864, 27.70263036,
164.08010452, 70.75066917, 98.44378876, 168.89543506,
129.46525513, 141.95611384, 85.71937762, 20.83727587,
114.74258617, 32.97733764, 46.94314617, 132.08748746,
34.28123094, 50.9155087 , 101.36663486, 61.49593031,
129.38305222, 143.80195002, 69.18963781, 29.8498935 ,
127.3139812 , 135.3309614 , 10.86515051, 24.39323345,
83.77082949, 19.40539599, 158.14371522, 153.7699104 ,
103.2520808 , 102.33671699, 28.07191471, 25.86542248,
69.64844548, 149.69146422, 92.47236736, 76.37524053,
85.06935039, 74.59726173, 17.34053295, 114.87770551,
42.12503406, 40.7773349 , 54.9953361 ])
Lasso model has the best r2score so far
Model 4: Support Vector Regression
# Building the best fit model using the gridsearch methodology
param_grid = {'C': [0.05, 0.1, 0.15],
'gamma': [1, 0.5],
'kernel': ['linear']}
grid = GridSearchCV(SVR(), param_grid, refit = True, verbose = 3)
# fitting the model for grid search
grid.fit(X_train_fc, y_train)
Fitting 5 folds for each of 6 candidates, totalling 30 fits
[CV 1/5] END ....C=0.05, gamma=1, kernel=linear;, score=0.616 total time= 0.0s
[CV 2/5] END ....C=0.05, gamma=1, kernel=linear;, score=0.435 total time= 0.0s
[CV 3/5] END ....C=0.05, gamma=1, kernel=linear;, score=0.551 total time= 0.0s
[CV 4/5] END ....C=0.05, gamma=1, kernel=linear;, score=0.483 total time= 0.0s
[CV 5/5] END ....C=0.05, gamma=1, kernel=linear;, score=0.560 total time= 0.0s
[CV 1/5] END ..C=0.05, gamma=0.5, kernel=linear;, score=0.616 total time= 0.0s
[CV 2/5] END ..C=0.05, gamma=0.5, kernel=linear;, score=0.435 total time= 0.0s
[CV 3/5] END ..C=0.05, gamma=0.5, kernel=linear;, score=0.551 total time= 0.0s
[CV 4/5] END ..C=0.05, gamma=0.5, kernel=linear;, score=0.483 total time= 0.0s
[CV 5/5] END ..C=0.05, gamma=0.5, kernel=linear;, score=0.560 total time= 0.0s
[CV 1/5] END .....C=0.1, gamma=1, kernel=linear;, score=0.853 total time= 0.0s
[CV 2/5] END .....C=0.1, gamma=1, kernel=linear;, score=0.606 total time= 0.0s
[CV 3/5] END .....C=0.1, gamma=1, kernel=linear;, score=0.732 total time= 0.0s
[CV 4/5] END .....C=0.1, gamma=1, kernel=linear;, score=0.718 total time= 0.0s
[CV 5/5] END .....C=0.1, gamma=1, kernel=linear;, score=0.777 total time= 0.0s
[CV 1/5] END ...C=0.1, gamma=0.5, kernel=linear;, score=0.853 total time= 0.0s
[CV 2/5] END ...C=0.1, gamma=0.5, kernel=linear;, score=0.606 total time= 0.0s
[CV 3/5] END ...C=0.1, gamma=0.5, kernel=linear;, score=0.732 total time= 0.0s
[CV 4/5] END ...C=0.1, gamma=0.5, kernel=linear;, score=0.718 total time= 0.0s
[CV 5/5] END ...C=0.1, gamma=0.5, kernel=linear;, score=0.777 total time= 0.0s
[CV 1/5] END ....C=0.15, gamma=1, kernel=linear;, score=0.888 total time= 0.0s
[CV 2/5] END ....C=0.15, gamma=1, kernel=linear;, score=0.688 total time= 0.0s
[CV 3/5] END ....C=0.15, gamma=1, kernel=linear;, score=0.787 total time= 0.0s
[CV 4/5] END ....C=0.15, gamma=1, kernel=linear;, score=0.790 total time= 0.0s
[CV 5/5] END ....C=0.15, gamma=1, kernel=linear;, score=0.831 total time= 0.0s
[CV 1/5] END ..C=0.15, gamma=0.5, kernel=linear;, score=0.888 total time= 0.0s
[CV 2/5] END ..C=0.15, gamma=0.5, kernel=linear;, score=0.688 total time= 0.0s
[CV 3/5] END ..C=0.15, gamma=0.5, kernel=linear;, score=0.787 total time= 0.0s
[CV 4/5] END ..C=0.15, gamma=0.5, kernel=linear;, score=0.790 total time= 0.0s
[CV 5/5] END ..C=0.15, gamma=0.5, kernel=linear;, score=0.831 total time= 0.0s
GridSearchCV(estimator=SVR(),
param_grid={'C': [0.05, 0.1, 0.15], 'gamma': [1, 0.5],
'kernel': ['linear']},
verbose=3)
# print best parameter after tuning
print('Best parameter after tuning:',grid.best_params_)
# print how our model looks after hyper-parameter tuning
print('How the model looks after hyper-param tuning:',grid.best_estimator_)
Best parameter after tuning: {'C': 0.15, 'gamma': 1, 'kernel': 'linear'}
How the model looks after hyper-param tuning: SVR(C=0.15, gamma=1, kernel='linear')
# Running the bestfit model and checking for r2score
svr = SVR(C=0.1, gamma=1, kernel='linear')
svr.fit(X_train, y_train)
svr_test_predict = svr.predict(X_test)
svr_r2_score = r2_score(y_test, svr_test_predict)
svr_r2_score
0.8643996577095221
Pickling The Model (Save and Load The Model)
with open('[4.4.1] SVR', 'wb') as f:
pickle.dump(svr, f)
with open('[4.4.1] SVR', 'rb') as f:
model_svr = pickle.load(f)
#testing the pickled model. reason: to make sure the pickle output showing the same as the current model (svr_test_predict)
model_svr.predict(X_test)
array([ 33.61462123, 164.36269696, 20.48650727, 38.68163826,
121.02233909, 19.33659598, 167.97054651, 79.71245358,
150.57426518, 172.92150154, 150.3639844 , 60.52480212,
100.72215544, 72.68408434, 112.71105755, 52.96940288,
85.7521067 , 35.48155735, 71.77059095, 175.68277946,
116.66901993, 176.59730809, 18.33681522, 28.50094336,
156.08413126, 82.25622977, 92.70473179, 163.03053081,
118.8752131 , 146.7946113 , 91.69587591, 11.80499872,
111.2579129 , 34.33433863, 45.4047336 , 140.40671253,
28.18633569, 52.28054402, 99.06751056, 55.60638826,
122.38383578, 137.39110587, 66.12547397, 28.39357435,
129.43458981, 133.33695526, 11.64593633, 14.30046499,
81.68968577, 21.39909846, 159.01439648, 149.47116859,
100.1127062 , 111.10998129, 27.66867627, 27.71575912,
71.34639659, 144.42100736, 82.47019504, 72.95081196,
79.75567094, 71.29616708, 11.62560707, 110.23471542,
27.18210951, 43.9198063 , 56.49803304])
# Training and testing the best fit model from the gridsearch
svr.fit(X_train_fc, y_train)
svr_test_predict1 = svr.predict(X_test_fc)
svr_r2fc_score1 = r2_score(y_test, svr_test_predict1)
svr_r2fc_score1
0.7837595066466638
Pickling The Model (Save and Load The Model)
with open('[4.4.2] SVR_Scaled Data', 'wb') as f:
pickle.dump(svr, f)
with open('[4.4.2] SVR_Scaled Data', 'rb') as f:
model_svr2 = pickle.load(f)
#testing the pickled model. reason: to make sure the pickle output showing the same as the current model (svr_test_predict1)
model_svr2.predict(X_test_fc)
array([ 51.16719319, 149.21590862, 36.54919052, 52.22075706,
111.10395449, 37.24018579, 139.16642421, 88.90701765,
134.96206797, 160.01278643, 139.91068987, 64.08340634,
107.66716247, 82.12050619, 107.27559541, 62.87839141,
93.57606035, 51.39241338, 80.45375634, 158.99354838,
109.94459273, 160.30012514, 38.04453935, 47.73957106,
144.36586558, 90.5598032 , 83.62541762, 150.82780178,
110.54001224, 122.37101352, 94.71267251, 34.92373102,
84.71230736, 46.47647793, 49.88835744, 123.70864577,
51.30375702, 73.62599846, 107.22059784, 56.10260556,
111.33221922, 136.38125732, 71.02105978, 41.1632985 ,
117.1462558 , 131.76020275, 32.12096076, 35.20918455,
82.77333167, 46.12570169, 139.07939918, 134.66435822,
100.93347795, 103.16115961, 41.42188225, 44.69021939,
78.76109663, 130.24174794, 89.65129204, 81.04970231,
83.51487819, 80.60313767, 30.85723387, 111.98974297,
43.42465048, 50.78456972, 62.6887925 ])
Remarks - SVR performs worst when scaled data is used, on the other hand SVR score in line with the linear regression models.
Model 5: PCA
First utilizing Eigen vectors and Eigen values to determine how many variables are contributing to the most amplitude of variance in the target.
# Creating the covariance matrix
X_std = sc.fit_transform(X_new)
transpose = X_std.T #transpose
transpose_df = pd.DataFrame(data=transpose) #transpose dataframe
cov_mat = np.cov(transpose)
cov_mat
array([[1.005 , 0.89446577, 0.80124336, 0.77819562, 0.8618311 ,
0.87091172, 0.68322998],
[0.89446577, 1.005 , 0.64158525, 0.90728581, 0.73433063,
0.89609901, 0.62270806],
[0.80124336, 0.64158525, 1.005 , 0.46590106, 0.83559335,
0.66615515, 0.6125262 ],
[0.77819562, 0.90728581, 0.46590106, 1.005 , 0.60945352,
0.8382459 , 0.52483638],
[0.8618311 , 0.73433063, 0.83559335, 0.60945352, 1.005 ,
0.75180265, 0.64395437],
[0.87091172, 0.89609901, 0.66615515, 0.8382459 , 0.75180265,
1.005 , 0.58424158],
[0.68322998, 0.62270806, 0.6125262 , 0.52483638, 0.64395437,
0.58424158, 1.005 ]])
#calculating the Eigen Vectors & Eigen Values
eig_vals, eig_vecs = np.linalg.eig(cov_mat) #get eigen_values and eigen_vectors - linalg = linear algebra
print('Eigen Vectors: \n' ,eig_vecs, '\n')
print('Eigen Values: \n' ,eig_vals)
Eigen Vectors:
[[ 0.41583476 0.03025878 -0.11326521 -0.51383018 -0.73608165 0.08639943
0.00586048]
[ 0.4029792 -0.32431067 0.01087487 0.76064565 -0.28943461 0.23837888
0.11460107]
[ 0.35227212 0.5376239 -0.34306535 -0.00257215 0.31276808 0.27910314
0.54170288]
[ 0.36275966 -0.56449326 0.08831211 -0.36784019 0.47267881 0.41424125
-0.10782523]
[ 0.38271598 0.33062425 -0.26306275 0.14470594 0.15349265 -0.08078736
-0.78992891]
[ 0.39692305 -0.25531078 -0.13338503 -0.03378907 0.16336849 -0.82117685
0.23940457]
[ 0.32400551 0.33424378 0.88009448 -0.00148721 0.05397246 -0.07320014
0.02148792]]
Eigen Values:
[5.38629931 0.7522166 0.45763728 0.05852459 0.09342933 0.13001594
0.15687695]
Interpretation: In order to decide which eigenvector(s) can dropped without losing an inordinate amount of information for the construction of lower-dimensional subspace, we require to inspect the corresponding eigenvalues: The eigenvectors with the lowest eigenvalues bear the least information about the distribution of the data; those are the ones can be dropped.
Decision: The prevalent approach is to rank the eigenvalues from highest to lowest in order choose the top k eigenvectors.
eig_pairs1 = dict(zip(eig_vals , eig_vecs)) # zipping two variables and converting to dictionary
eigen_array = list(eig_pairs1.items()) #converting to list
eigen_array = np.array(eigen_array) #converting to array
print('Sort Eigen Values & Eigen Vectors: \n',eigen_array)
Sort Eigen Values & Eigen Vectors:
[[5.386299306379085
array([ 0.41583476, 0.03025878, -0.11326521, -0.51383018, -0.73608165,
0.08639943, 0.00586048]) ]
[0.7522165981994982
array([ 0.4029792 , -0.32431067, 0.01087487, 0.76064565, -0.28943461,
0.23837888, 0.11460107]) ]
[0.4576372801011403
array([ 0.35227212, 0.5376239 , -0.34306535, -0.00257215, 0.31276808,
0.27910314, 0.54170288]) ]
[0.05852459409182731
array([ 0.36275966, -0.56449326, 0.08831211, -0.36784019, 0.47267881,
0.41424125, -0.10782523]) ]
[0.0934293311702608
array([ 0.38271598, 0.33062425, -0.26306275, 0.14470594, 0.15349265,
-0.08078736, -0.78992891]) ]
[0.13001593855227458
array([ 0.39692305, -0.25531078, -0.13338503, -0.03378907, 0.16336849,
-0.82117685, 0.23940457]) ]
[0.15687695150591968
array([ 0.32400551, 0.33424378, 0.88009448, -0.00148721, 0.05397246,
-0.07320014, 0.02148792]) ]]
Finding the Explained Variance which tells us how much information (variance) can be attributed to each of the principal components
tot = sum(eig_vals)
var_exp = [(i / tot)*100 for i in sorted(eig_vals, reverse=True)] #reverse is true means arranging in the descending order
print('Explained Variance:')
var_exp
Explained Variance:
[76.564311391316,
10.692488958059666,
6.505149681608244,
2.229949559430271,
1.8481299012405754,
1.32806440895893,
0.8319060993863151]
Interpretation: above analysis shows that two vectors contribute up to > 87% of the variance in the target variable.
Decision: we run the PCA analysis and safely reduce the no of features/dimensions to 2 to predict the target accurately
pca = PCA(n_components=2)
principalComponents = pca.fit_transform(X_new)
principalDataframe = pd.DataFrame(data = principalComponents, columns = ['PC1', 'PC2'])
#adding y to the data set to visualize the new dataset
newDataframe = pd.concat([principalDataframe, y],axis = 1)
newDataframe.head()
| PC1 | PC2 | price | |
|---|---|---|---|
| 0 | 0.857629 | 25.745987 | 117 |
| 1 | 0.857629 | 25.745987 | 136 |
| 2 | 27.756635 | 28.303743 | 136 |
| 3 | -10.014828 | -12.083144 | 122 |
| 4 | 36.226517 | 2.680239 | 147 |
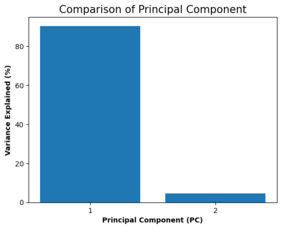
#checking the % of variance in price explained by the 2 components
percent_variance = np.round(pca.explained_variance_ratio_* 100, decimals =2)
columns = ['1', '2']
plt.bar(x= range(1,3), height=percent_variance, tick_label=columns)
plt.ylabel('Variance Explained (%)',fontweight='bold')
plt.xlabel('Principal Component (PC)',fontweight='bold')
plt.title('Comparison of Principal Component', size = 15)
plt.savefig('[Fig 4.5.1] PCA_Comparison of Principal Component.png', dpi = 300)
plt.show()
#scatter plot
plt.scatter(principalDataframe.PC1, principalDataframe.PC2)
plt.title('PC1 against PC2',size = 15)
plt.xlabel('PC1')
plt.ylabel('PC2')
plt.savefig('[Fig 4.5.2] PCA_PC1 against PC2 Plot.png', dpi = 300)
plt.show()
No significant pattern
pca.explained_variance_ratio_
array([0.90400936, 0.04652694])
Interpretation: These values show that the first principal component PC1 explains most of the variance in the target.
Decision: Build and visualizing the PCA model further. We will first disunite the train and test data and do the prediction.
def pca_dec(data, n):
pca = PCA(n)
princi_components = pca.fit_transform(data)
return princi_components, pca
#Decomposing the train set:
X_trn_pca, pca_train = pca_dec(X_trn_new, 2)
#Decomposing the test set:
X_tst_pca, pca_test = pca_dec(X_tst_new, 2)
X_trn_df = pd.DataFrame(data = X_trn_pca, columns = ['X_trn_PC1', 'X_trn_PC2'])
print(X_trn_df.head())
X_tst_df = pd.DataFrame(data = X_tst_pca, columns = ['X_tst_PC1', 'X_tst_PC2'])
print(X_tst_df.head())
X_trn_PC1 X_trn_PC2
0 49.980616 -9.744192
1 -61.296918 -7.668622
2 -47.127075 12.557807
3 -75.490579 -4.750386
4 3.995841 -0.153073
X_tst_PC1 X_tst_PC2
0 -63.997451 -0.032976
1 92.727444 6.319053
2 -88.593318 9.013806
3 -47.712789 -1.474157
4 27.551259 -8.689390
#fitting the model
lm.fit(X_trn_df,y_trn_new)
# predicting the test data
y_pred_pca = lm.predict(X_tst_df)
# checking the R2 score
linear_PCA_score = r2_score(y_tst_new,y_pred_pca)
linear_PCA_score
0.8638450042408081
Pickling The Model (Save and Load The Model)
with open('[4.5.1] PCA', 'wb') as f:
pickle.dump(lm, f)
with open('[4.5.1] PCA', 'rb') as f:
model_pca = pickle.load(f)
#testing the pickled model. reason: to make sure the pickle output showing the same as the current model (y_pred_pca)
model_pca.predict(X_tst_df)
array([ 37.9645029 , 172.52744095, 21.21491852, 51.03253584,
111.1889147 , 25.38711314, 173.82916435, 88.80386874,
146.8090489 , 182.55372327, 166.16350822, 66.68738375,
112.55506802, 79.55414464, 110.4527255 , 55.12354572,
84.5658018 , 46.42722245, 81.73823254, 174.46440435,
136.25278015, 175.67203409, 36.90366031, 35.08919826,
170.70061726, 86.69380947, 90.77482424, 171.98816234,
139.32298908, 131.17284191, 87.39179272, 21.05818118,
101.22975087, 35.43966643, 61.7648987 , 133.13557043,
52.79809295, 58.33223183, 110.25241132, 57.8805349 ,
135.62780735, 143.60858845, 73.42031937, 33.8301877 ,
131.5718391 , 137.31360752, 16.4349561 , 26.01920097,
93.35467871, 24.34346701, 153.7550102 , 145.27394444,
101.56800115, 109.65780716, 31.527531 , 26.22903964,
82.97023874, 155.55407388, 92.6416299 , 78.87094644,
89.52036816, 76.56828975, 16.95296254, 123.42276321,
40.21871637, 51.70121555, 75.64549286])
#Creating a table with the explained variance ratio for the train data set
names_pcas = [f"PCA Component {i}" for i in range(1, 3, 1)]
scree = pd.DataFrame(list(zip(names_pcas, pca_train.explained_variance_ratio_)), columns=["Component", "Explained Variance Ratio"])
print(scree)
Component Explained Variance Ratio
0 PCA Component 1 0.896766
1 PCA Component 2 0.048400
Interpretation: Based on the above it is conspicuous that PCA1 of the training dataset has availed expound virtually 90% of the variance in price (target), but we additionally need to find which of the features from the original dataset make up each of the components and which features contribute the highest to that eigenvectors.
Decision: Sorting the values of the first principal component by how large each one and sorting the absolute values of the first principal component by magnitude
#Sorting the values of the first principal component by how large each one is
df = pd.DataFrame({'PCA':pca_train.components_[0], 'Variable Names':list(X_trn_new.columns)})
df = df.sort_values('PCA', ascending=False)
#Sorting the absolute values of the first principal component by magnitude
df2 = pd.DataFrame(df)
df2['PCA']=df2['PCA'].apply(np.absolute)
df2 = df2.sort_values('PCA', ascending=False)
#print(df2['Variable Names'][0:11])
df.head()
| PCA | Variable Names | |
|---|---|---|
| 0 | 0.859986 | curb_weight |
| 1 | 0.329952 | length |
| 2 | 0.208613 | horsepower |
| 3 | 0.198711 | wheel_base |
| 5 | 0.166743 | width |
Interpretation: curb_weight is the most descriptive factor in PCA1 and contributes about 86% to PCA1, this makes an abundance of sense intuitively as the higher the curb weight the higher the other values are like length, width etc and hence is a significant factor in determining the price.
Model 6 - Final Iteration to Find Tune The PCA (Linear Regression Combo Model)
by using only those variables in the Linear Regression Model that make up the PCA1 components from above PCA Analysis.
# creating a data set to include only the components of the PCA1 above
X_PCA = x[['curb_weight', 'length','wheel_base','horsepower', 'width']].copy()
# training and splitting the data set
X_trn_PCA, X_tst_PCA, y_trn_PCA, y_tst_PCA = train_test_split(X_PCA, y, test_size=0.33, random_state=42)
print(X_trn_PCA.shape, X_tst_PCA.shape, y_trn_PCA.shape, y_tst_PCA.shape)
# scaling the data set
X_trn_PCA_fc = sc.fit_transform(X_trn_PCA)
X_tst_PCA_fc = sc.transform(X_tst_PCA)
#fitting the model
lm.fit(X_trn_PCA_fc,y_trn_PCA)
# predicting the test data
y_pred_PCA = lm.predict(X_tst_PCA_fc)
# checking the R2 score
linear_FinalRegrPCA_score = r2_score(y_tst_PCA,y_pred_PCA)
linear_FinalRegrPCA_score
(134, 5) (67, 5) (134,) (67,)
0.8812197638869728
Interpretation: The r2score has improved by using only the variables that make up the PCA1 component which betokens that these are the most consequential predictor variables to be considered for the car price prediction.
Pickling The Model (Save and Load The Model)
with open('[4.5.2] PCA (Linear Regression Combo Model)', 'wb') as f:
pickle.dump(lm, f)
with open('[4.5.2] PCA (Linear Regression Combo Model)', 'rb') as f:
model_pca = pickle.load(f)
#testing the pickled model. reason: to make sure the pickle output showing the same as the current model (y_pred_PCA)
model_pca.predict(X_tst_PCA_fc)
array([ 30.74360127, 173.57112374, 21.74652627, 43.45626682,
112.35831945, 21.61600579, 169.22717335, 89.23820946,
153.42475431, 183.79120412, 159.93260049, 59.56664013,
108.70381117, 73.36807852, 112.60980703, 46.24767767,
81.93197674, 40.43567559, 74.43145855, 178.32923086,
132.54734325, 181.8906238 , 27.03987554, 29.13324515,
164.02119982, 75.95501644, 95.23767692, 169.33167933,
134.56107074, 141.03022976, 86.53727535, 19.16285949,
106.53860934, 30.50239658, 46.66850385, 128.32143598,
38.30262547, 55.88322274, 107.19351556, 57.20532959,
131.41269502, 145.88414287, 71.28145647, 27.15375637,
129.04277478, 137.20080521, 11.23867913, 23.52419749,
85.60234386, 22.08519895, 154.34938341, 152.41789056,
103.85851436, 107.57552102, 25.64346075, 24.46121413,
71.94949838, 150.96712485, 91.75536882, 78.01481834,
83.26124916, 76.50452273, 14.49799789, 116.88844137,
36.51338227, 40.63077699, 58.65781791])
Summary of Analysis
# Summary of the Accuracy scores for test data
model_ev = pd.DataFrame({'Model': ['Linear Regression','Ridge Regression','Lasso Regression','SVR',
'PCA', 'PCA-Linear Combo'], 'r2 score (%)': [round(linear_FinalRegr_score*100, 3),
round(Ridge_RegrLn_Score*100, 3), round(Lasso_RegrLn_Score*100, 3),round(svr_r2_score*100, 3),round(linear_PCA_score*100, 3), round(linear_FinalRegrPCA_score*100, 3)]})
model_ev = model_ev.sort_values('r2 score (%)', ascending=False)
model_ev
| Model | r2 score (%) | |
|---|---|---|
| 5 | PCA-Linear Combo | 88.122 |
| 2 | Lasso Regression | 87.990 |
| 0 | Linear Regression | 87.983 |
| 1 | Ridge Regression | 87.983 |
| 3 | SVR | 86.440 |
| 4 | PCA | 86.385 |
#Bar plots of the r2scores for visualization
colors = ['red','green','blue','c','orange', 'yellow']
plt.figure(figsize=(15,7))
plt.title("Barplot of 6 Models", size = 15)
plt.xlabel("Models")
plt.xticks(rotation=0)
plt.ylim(86,88.5, 0.001)
plt.ylabel("r2 scores")
plt.bar(model_ev['Model'],model_ev['r2 score (%)'],color = colors)
plt.savefig('[Fig 5] Performance of Each Models', dpi = 300)
plt.show()
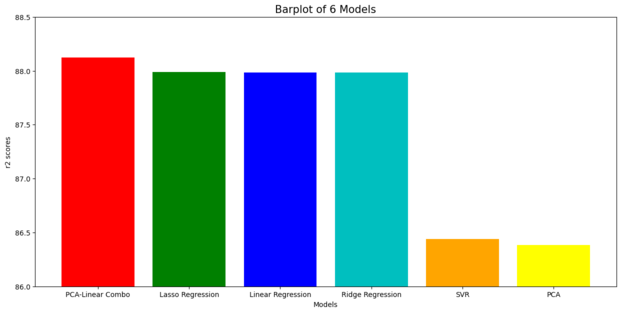
The most prominent model falls on PCA-Linear Regression Combo Model
Lasso - Ridge Regression models look identically tantamount (predicated on the graph above). However, speaking about the details, the digit precision of each r2 score needs to be considered.Conclusion: PCA-Linear Regression Combo Model, it indicates better performance for predicting target (price).
Recommendation:
- Lasso regression model is one of the best modeling techinque for this project. It also takes care of the multicollinearity issue that is present in this dataset without having to do any transformation of the dataset. It also takes less time in running compared to SVR.
- The PCA Analysis took care of most of the multicollinearity issues and provided us 5 predictor variables (curb weight, length, horse power, wheel base and width) that can accurately predict the car prices. The Linear Regression model with these 5 predictor variables gave the best r2score and so we recommend the usage of this PCA-Linear Regression Model for this project.
Project Risks
We have taken the 10 best features from the feature selection algorithm to reduce the dimensionality problem for modeling purposes, so if there are any interaction effects between the features that have not been modeled, due to lack of expertise in this field, there may be considerable amount of underperformance in the model when it is implemented.
The project postulates that the dataset utilized for modeling in this project is a representative of the population dataset else the models may not provide the accuracies that are shown here.