Project Team ID = PTID-CDS-JUL21-1171 (Members - Diana, Hema, Pavithra and Sophiya)
Project ID = PRCP-1009-CellphonePrice (Cellphone Price Range)
Phase 1: Loading the Dataset and Understanding The Difference Features and Target
import numpy as np
%matplotlib inline
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
data = pd.read_csv('datasets_11167_15520_train.csv')
data.head()
| battery_power | blue | clock_speed | dual_sim | fc | four_g | int_memory | m_dep | mobile_wt | n_cores | ... | px_height | px_width | ram | sc_h | sc_w | talk_time | three_g | touch_screen | wifi | price_range | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 842 | 0 | 2.2 | 0 | 1 | 0 | 7 | 0.6 | 188 | 2 | ... | 20 | 756 | 2549 | 9 | 7 | 19 | 0 | 0 | 1 | 1 |
| 1 | 1021 | 1 | 0.5 | 1 | 0 | 1 | 53 | 0.7 | 136 | 3 | ... | 905 | 1988 | 2631 | 17 | 3 | 7 | 1 | 1 | 0 | 2 |
| 2 | 563 | 1 | 0.5 | 1 | 2 | 1 | 41 | 0.9 | 145 | 5 | ... | 1263 | 1716 | 2603 | 11 | 2 | 9 | 1 | 1 | 0 | 2 |
| 3 | 615 | 1 | 2.5 | 0 | 0 | 0 | 10 | 0.8 | 131 | 6 | ... | 1216 | 1786 | 2769 | 16 | 8 | 11 | 1 | 0 | 0 | 2 |
| 4 | 1821 | 1 | 1.2 | 0 | 13 | 1 | 44 | 0.6 | 141 | 2 | ... | 1208 | 1212 | 1411 | 8 | 2 | 15 | 1 | 1 | 0 | 1 |
5 rows × 21 columns
Description of the Dataset
battery_power:Total energy a battery can store in one time measured in mAh
blue:Has bluetooth or not
clock_speed:speed at which microprocessor executes instructions
dual_sim:Has dual sim support or not
fc:Front Camera mega pixels
four_g:Has 4G or not
int_memory:Internal Memory in Gigabytes
m_dep:Mobile Depth in cm
mobile_wt:Weight of mobile phone
n_cores:Number of cores of processor
pc:Primary Camera mega pixels
px_height:Pixel Resolution Height
px_width:Pixel Resolution Width
ram:Random Access Memory in Megabytes
sc_h:Screen Height of mobile in cm
sc_w:Screen Width of mobile in cm
talk_time:longest time that a single battery charge will last when you are
three_g:Has 3G or not
touch_screen:Has touch screen or not
wifi:Has wifi or not
price_range: three classifications could be high range, mid range and low range (dont know how the target is scaled)
print("____________________________________________")
print(" Info of The Dataset")
print("____________________________________________")
data.info()
____________________________________________
Info of The Dataset
____________________________________________
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 2000 entries, 0 to 1999
Data columns (total 21 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 battery_power 2000 non-null int64
1 blue 2000 non-null int64
2 clock_speed 2000 non-null float64
3 dual_sim 2000 non-null int64
4 fc 2000 non-null int64
5 four_g 2000 non-null int64
6 int_memory 2000 non-null int64
7 m_dep 2000 non-null float64
8 mobile_wt 2000 non-null int64
9 n_cores 2000 non-null int64
10 pc 2000 non-null int64
11 px_height 2000 non-null int64
12 px_width 2000 non-null int64
13 ram 2000 non-null int64
14 sc_h 2000 non-null int64
15 sc_w 2000 non-null int64
16 talk_time 2000 non-null int64
17 three_g 2000 non-null int64
18 touch_screen 2000 non-null int64
19 wifi 2000 non-null int64
20 price_range 2000 non-null int64
dtypes: float64(2), int64(19)
memory usage: 328.2 KB
data.isnull().sum()
battery_power 0
blue 0
clock_speed 0
dual_sim 0
fc 0
four_g 0
int_memory 0
m_dep 0
mobile_wt 0
n_cores 0
pc 0
px_height 0
px_width 0
ram 0
sc_h 0
sc_w 0
talk_time 0
three_g 0
touch_screen 0
wifi 0
price_range 0
dtype: int64
data.describe().T
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| battery_power | 2000.0 | 1238.51850 | 439.418206 | 501.0 | 851.75 | 1226.0 | 1615.25 | 1998.0 |
| blue | 2000.0 | 0.49500 | 0.500100 | 0.0 | 0.00 | 0.0 | 1.00 | 1.0 |
| clock_speed | 2000.0 | 1.52225 | 0.816004 | 0.5 | 0.70 | 1.5 | 2.20 | 3.0 |
| dual_sim | 2000.0 | 0.50950 | 0.500035 | 0.0 | 0.00 | 1.0 | 1.00 | 1.0 |
| fc | 2000.0 | 4.30950 | 4.341444 | 0.0 | 1.00 | 3.0 | 7.00 | 19.0 |
| four_g | 2000.0 | 0.52150 | 0.499662 | 0.0 | 0.00 | 1.0 | 1.00 | 1.0 |
| int_memory | 2000.0 | 32.04650 | 18.145715 | 2.0 | 16.00 | 32.0 | 48.00 | 64.0 |
| m_dep | 2000.0 | 0.50175 | 0.288416 | 0.1 | 0.20 | 0.5 | 0.80 | 1.0 |
| mobile_wt | 2000.0 | 140.24900 | 35.399655 | 80.0 | 109.00 | 141.0 | 170.00 | 200.0 |
| n_cores | 2000.0 | 4.52050 | 2.287837 | 1.0 | 3.00 | 4.0 | 7.00 | 8.0 |
| pc | 2000.0 | 9.91650 | 6.064315 | 0.0 | 5.00 | 10.0 | 15.00 | 20.0 |
| px_height | 2000.0 | 645.10800 | 443.780811 | 0.0 | 282.75 | 564.0 | 947.25 | 1960.0 |
| px_width | 2000.0 | 1251.51550 | 432.199447 | 500.0 | 874.75 | 1247.0 | 1633.00 | 1998.0 |
| ram | 2000.0 | 2124.21300 | 1084.732044 | 256.0 | 1207.50 | 2146.5 | 3064.50 | 3998.0 |
| sc_h | 2000.0 | 12.30650 | 4.213245 | 5.0 | 9.00 | 12.0 | 16.00 | 19.0 |
| sc_w | 2000.0 | 5.76700 | 4.356398 | 0.0 | 2.00 | 5.0 | 9.00 | 18.0 |
| talk_time | 2000.0 | 11.01100 | 5.463955 | 2.0 | 6.00 | 11.0 | 16.00 | 20.0 |
| three_g | 2000.0 | 0.76150 | 0.426273 | 0.0 | 1.00 | 1.0 | 1.00 | 1.0 |
| touch_screen | 2000.0 | 0.50300 | 0.500116 | 0.0 | 0.00 | 1.0 | 1.00 | 1.0 |
| wifi | 2000.0 | 0.50700 | 0.500076 | 0.0 | 0.00 | 1.0 | 1.00 | 1.0 |
| price_range | 2000.0 | 1.50000 | 1.118314 | 0.0 | 0.75 | 1.5 | 2.25 | 3.0 |
data['price_range'].value_counts()
0 500
1 500
2 500
3 500
Name: price_range, dtype: int64
Teams Remarks: Data seems to be balanced for each of the classes with no null values for any of the variables
Phase 2: Data Preparation
from sklearn.preprocessing import LabelEncoder
# ENCODING THE ENTIRE DATA SET
label = LabelEncoder()
data1 = data.apply(label.fit_transform)
data1
| battery_power | blue | clock_speed | dual_sim | fc | four_g | int_memory | m_dep | mobile_wt | n_cores | ... | px_height | px_width | ram | sc_h | sc_w | talk_time | three_g | touch_screen | wifi | price_range | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 255 | 0 | 17 | 0 | 1 | 0 | 5 | 5 | 108 | 1 | ... | 18 | 186 | 963 | 4 | 7 | 17 | 0 | 0 | 1 | 1 |
| 1 | 382 | 1 | 0 | 1 | 0 | 1 | 51 | 6 | 56 | 2 | ... | 738 | 1099 | 998 | 12 | 3 | 5 | 1 | 1 | 0 | 2 |
| 2 | 48 | 1 | 0 | 1 | 2 | 1 | 39 | 8 | 65 | 4 | ... | 962 | 901 | 984 | 6 | 2 | 7 | 1 | 1 | 0 | 2 |
| 3 | 88 | 1 | 20 | 0 | 0 | 0 | 8 | 7 | 51 | 5 | ... | 935 | 954 | 1055 | 11 | 8 | 9 | 1 | 0 | 0 | 2 |
| 4 | 954 | 1 | 7 | 0 | 13 | 1 | 42 | 5 | 61 | 1 | ... | 928 | 517 | 499 | 3 | 2 | 13 | 1 | 1 | 0 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1995 | 216 | 1 | 0 | 1 | 0 | 1 | 0 | 7 | 26 | 5 | ... | 939 | 1022 | 180 | 8 | 4 | 17 | 1 | 1 | 0 | 0 |
| 1996 | 1064 | 1 | 21 | 1 | 0 | 0 | 37 | 1 | 107 | 3 | ... | 745 | 1080 | 732 | 6 | 10 | 14 | 1 | 1 | 1 | 2 |
| 1997 | 1027 | 0 | 4 | 1 | 1 | 1 | 34 | 6 | 28 | 7 | ... | 716 | 834 | 1158 | 4 | 1 | 3 | 1 | 1 | 0 | 3 |
| 1998 | 732 | 0 | 4 | 0 | 4 | 1 | 44 | 0 | 65 | 4 | ... | 283 | 120 | 266 | 13 | 10 | 17 | 1 | 1 | 1 | 0 |
| 1999 | 8 | 1 | 15 | 1 | 5 | 1 | 43 | 8 | 88 | 5 | ... | 413 | 184 | 1526 | 14 | 4 | 0 | 1 | 1 | 1 | 3 |
2000 rows × 21 columns
# splitting the data into X and Y so we can do feature selection
x = data1.drop('price_range', axis=1)
y = data1['price_range']
print(x.head())
y.head()
battery_power blue clock_speed dual_sim fc four_g int_memory m_dep \
0 255 0 17 0 1 0 5 5
1 382 1 0 1 0 1 51 6
2 48 1 0 1 2 1 39 8
3 88 1 20 0 0 0 8 7
4 954 1 7 0 13 1 42 5
mobile_wt n_cores pc px_height px_width ram sc_h sc_w talk_time \
0 108 1 2 18 186 963 4 7 17
1 56 2 6 738 1099 998 12 3 5
2 65 4 6 962 901 984 6 2 7
3 51 5 9 935 954 1055 11 8 9
4 61 1 14 928 517 499 3 2 13
three_g touch_screen wifi
0 0 0 1
1 1 1 0
2 1 1 0
3 1 0 0
4 1 1 0
0 1
1 2
2 2
3 2
4 1
Name: price_range, dtype: int64
Too much input variables will be decreasing the accuracy of models so using the feature selection technique to reduce the no. of input variables that contribute less to predicting the target
# FEATURE SELECTION TECHNIQUE
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
#apply SelectKBest class to extract top 10 best features
bestfeatures = SelectKBest(score_func=chi2, k=10)
fit = bestfeatures.fit(x,y)
dfscores = pd.DataFrame(fit.scores_)
dfcolumns = pd.DataFrame(x.columns)
#concat two dataframes for better visualization
featurescores = pd.concat([dfcolumns,dfscores], axis=1)
featurescores.columns = ['Specs', 'Score'] #naming the data
print(featurescores.nlargest(10, 'Score'))
Specs Score
13 ram 428852.649505
0 battery_power 16835.196948
12 px_width 12243.820689
11 px_height 9989.550625
8 mobile_wt 223.407824
6 int_memory 95.819130
15 sc_w 16.480319
14 sc_h 16.194553
16 talk_time 16.174231
9 n_cores 11.681721
The top 10 features are now used to build the models for comparison
Modifying the dataset to include only these 10 important features
X = x[['ram', 'battery_power', 'px_width','px_height','mobile_wt','int_memory','sc_w','sc_h','talk_time','n_cores']].copy()
X.head()
| ram | battery_power | px_width | px_height | mobile_wt | int_memory | sc_w | sc_h | talk_time | n_cores | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 963 | 255 | 186 | 18 | 108 | 5 | 7 | 4 | 17 | 1 |
| 1 | 998 | 382 | 1099 | 738 | 56 | 51 | 3 | 12 | 5 | 2 |
| 2 | 984 | 48 | 901 | 962 | 65 | 39 | 2 | 6 | 7 | 4 |
| 3 | 1055 | 88 | 954 | 935 | 51 | 8 | 8 | 11 | 9 | 5 |
| 4 | 499 | 954 | 517 | 928 | 61 | 42 | 2 | 3 | 13 | 1 |
Phase 3: Exploratory Data Analysis
# Checking for the skewness and kurtosis for 10 selected features and target
print("Skewness of ram: %f" %X['ram'].skew())
print("Kurtosis of ram: %f" %X['ram'].kurt())
print("Skewness of battery power: %f" %X['battery_power'].skew())
print("Kurtosis of battery power: %f" %X['battery_power'].kurt())
print("Skewness of Pixel Width: %f" %X['px_width'].skew())
print("Kurtosis of Pixel Width: %f" %X['px_width'].kurt())
print("Skewness of Pixel Height: %f" %X['px_height'].skew())
print("Kurtosis of Pixel Height: %f" %X['px_height'].kurt())
print("Skewness of Mobile Weight: %f" %X['mobile_wt'].skew())
print("Kurtosis of Mobile Weight: %f" %X['mobile_wt'].kurt())
print("Skewness of Internal Memory: %f" %X['int_memory'].skew())
print("Kurtosis of Internal Memory: %f" %X['int_memory'].kurt())
print("Skewness of Screen Width: %f" %X['sc_w'].skew())
print("Kurtosis of Screen Width: %f" %X['sc_w'].kurt())
print("Skewness of Screen Height: %f" %X['sc_h'].skew())
print("Kurtosis of Screen Height: %f" %X['sc_h'].kurt())
print("Skewness of Talking Time Performance: %f" %X['talk_time'].skew())
print("Kurtosis of Talking Time Performance: %f" %X['talk_time'].kurt())
print("Skewness of Number Cores Processor: %f" %X['n_cores'].skew())
print("Kurtosis of Number Cores Processor: %f" %X['n_cores'].kurt())
print("Skewness of Price Range: %f" %data1['price_range'].skew())
print("Kurtosis of Price Range: %f" %data1['price_range'].kurt())
Skewness of ram: -0.001103
Kurtosis of ram: -1.171483
Skewness of battery power: 0.028115
Kurtosis of battery power: -1.206078
Skewness of Pixel Width: 0.029001
Kurtosis of Pixel Width: -1.198770
Skewness of Pixel Height: 0.237204
Kurtosis of Pixel Height: -1.090681
Skewness of Mobile Weight: 0.006558
Kurtosis of Mobile Weight: -1.210376
Skewness of Internal Memory: 0.057889
Kurtosis of Internal Memory: -1.216074
Skewness of Screen Width: 0.633787
Kurtosis of Screen Width: -0.389523
Skewness of Screen Height: -0.098884
Kurtosis of Screen Height: -1.190791
Skewness of Talking Time Performance: 0.009512
Kurtosis of Talking Time Performance: -1.218591
Skewness of Number Cores Processor: 0.003628
Kurtosis of Number Cores Processor: -1.229750
Skewness of Price Range: 0.000000
Kurtosis of Price Range: -1.360400
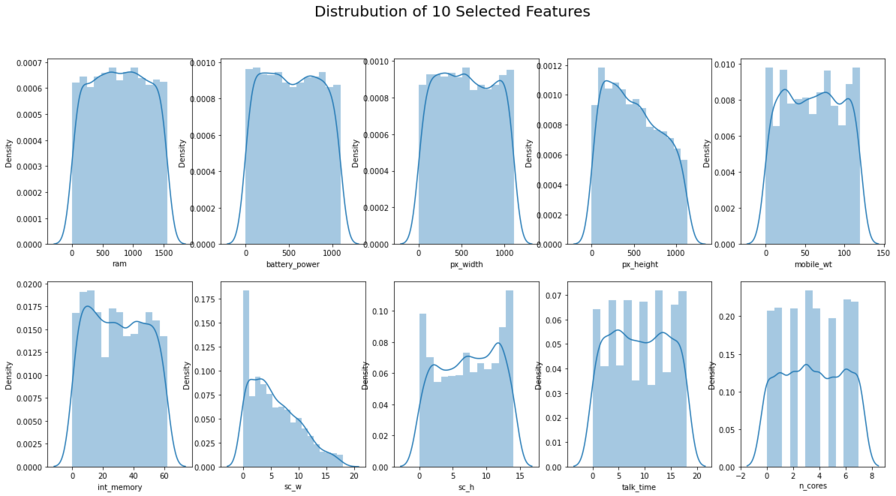
Reference - Skewness between -0.5 and 0.5: data fairly symmetrical. Skewness between -1 and – 0.5 or between 0.5 and 1: data moderately skewed. Skewness is less than -1 or greater than 1: the data are highly skewed. Kurtosis bettween -2 and +2 are considered acceptable.
Teams Remarks: The Skewness and Kurtosis for all the predictor and target variables falls within the acceptable range.
figure, ax = plt.subplots(2,5, figsize=(20,10))
#See the distrubution of the data
plt.suptitle('Distrubution of 10 Selected Features', size = 20)
sns.distplot(data1['ram'],ax=ax[0,0])
sns.distplot(data1['battery_power'], ax=ax[0,1])
sns.distplot(data1['px_width'], ax=ax[0,2])
sns.distplot(data1['px_height'], ax=ax[0,3])
sns.distplot(data1['mobile_wt'], ax=ax[0,4])
sns.distplot(data1['int_memory'], ax=ax[1,0])
sns.distplot(data1['sc_w'], ax=ax[1,1])
sns.distplot(data1['sc_h'], ax=ax[1,2])
sns.distplot(data1['talk_time'], ax=ax[1,3])
sns.distplot(data1['n_cores'], ax=ax[1,4])
plt.savefig('EDA - Distplot Visualization.png', dpi = 300)
plt.show()
#slicing 10 selected features to display boxplot. reasons to do that: the range of some features are different
slice_X1 = X[['ram', 'battery_power', 'px_width', 'px_height', 'mobile_wt']]
slice_X1
slice_X2 = X[['int_memory', 'sc_w', 'sc_h', 'talk_time', 'n_cores']]
slice_X2
| int_memory | sc_w | sc_h | talk_time | n_cores | |
|---|---|---|---|---|---|
| 0 | 5 | 7 | 4 | 17 | 1 |
| 1 | 51 | 3 | 12 | 5 | 2 |
| 2 | 39 | 2 | 6 | 7 | 4 |
| 3 | 8 | 8 | 11 | 9 | 5 |
| 4 | 42 | 2 | 3 | 13 | 1 |
| ... | ... | ... | ... | ... | ... |
| 1995 | 0 | 4 | 8 | 17 | 5 |
| 1996 | 37 | 10 | 6 | 14 | 3 |
| 1997 | 34 | 1 | 4 | 3 | 7 |
| 1998 | 44 | 10 | 13 | 17 | 4 |
| 1999 | 43 | 4 | 14 | 0 | 5 |
2000 rows × 5 columns
#Boxplot to check any outlier
plt.figure(figsize=(20,15))
plt.subplot(2, 2, 1)
sns.set_theme(style="darkgrid")
ax = sns.boxplot(data=slice_X1, orient="w", palette="Dark2")
plt.subplot(2, 2, 2)
sns.set_theme(style="darkgrid")
ax = sns.boxplot(data=slice_X2, orient="w", palette="Dark2_r")
plt.suptitle("Boxplot of of 10 Selected Features", size = 25)
plt.savefig('EDA - Boxplot Visualization.png', dpi = 300)
plt.show()
print('Checking...========================================\nResult: Outliers Not Found')
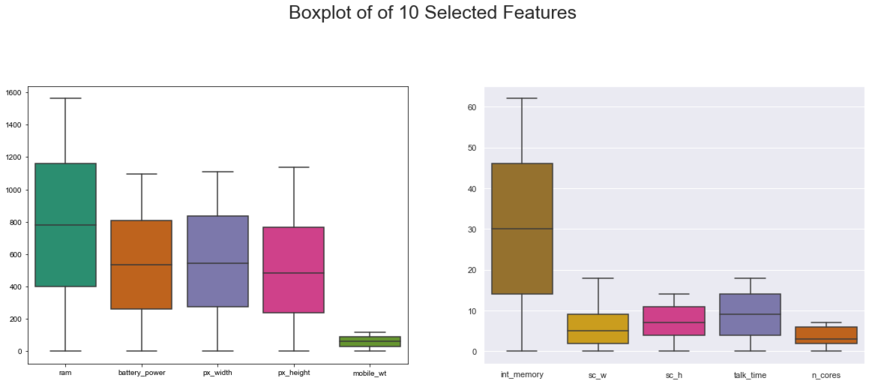
Checking...========================================
Result: Outliers Not Found
No significant outliers in the dataset
#combining the x (features) and y (target) to show the heatmap and pairplot analysis
xydata = data1[['ram', 'battery_power', 'px_width','px_height','mobile_wt','int_memory','sc_w','sc_h','talk_time','n_cores', 'price_range']].copy()
xydata.head()
| ram | battery_power | px_width | px_height | mobile_wt | int_memory | sc_w | sc_h | talk_time | n_cores | price_range | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 963 | 255 | 186 | 18 | 108 | 5 | 7 | 4 | 17 | 1 | 1 |
| 1 | 998 | 382 | 1099 | 738 | 56 | 51 | 3 | 12 | 5 | 2 | 2 |
| 2 | 984 | 48 | 901 | 962 | 65 | 39 | 2 | 6 | 7 | 4 | 2 |
| 3 | 1055 | 88 | 954 | 935 | 51 | 8 | 8 | 11 | 9 | 5 | 2 |
| 4 | 499 | 954 | 517 | 928 | 61 | 42 | 2 | 3 | 13 | 1 | 1 |
#Heatmap to shows the correlation
plt.figure(figsize=(20,15))
sns.heatmap(xydata.corr(),cmap='nipy_spectral',annot=True)
plt.title('Heatmap Shows The Relationship (correlation) Between Selected Features',
fontsize=25)
plt.savefig('EDA - Heatmap Visualization.png', dpi = 300)
plt.show()
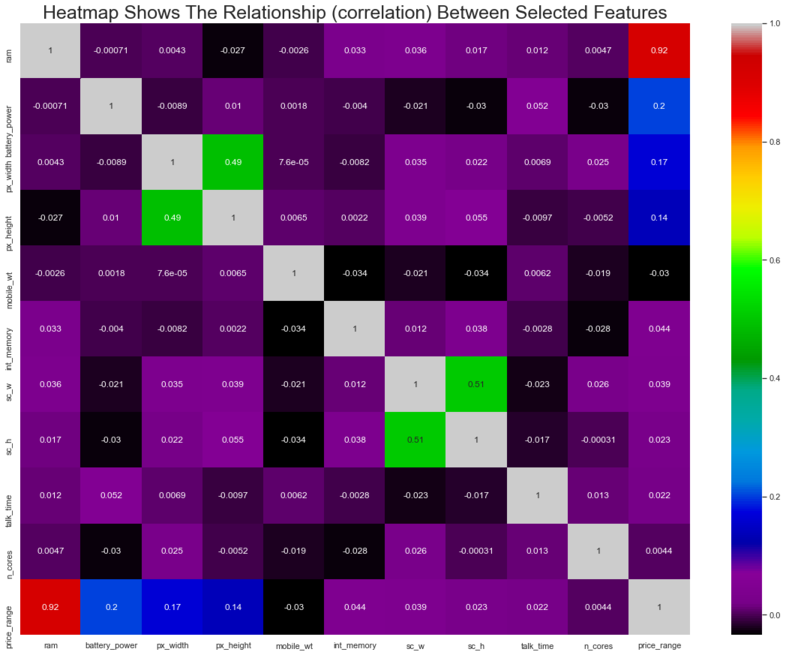
Teams Remarks: Ram dictates 92% of the price range classification. Screen width and screen height seem to be correlated, so we may merge them to reduce any dimensionality problems. For now we have decided to leave it in there for modelling purposes. No other major collinearlity issues
#pairplot analysis
sns.pairplot(xydata, hue = 'price_range', corner=True, palette='gnuplot')
plt.savefig('EDA - Pairplot Visualization.png', dpi = 300)
plt.show()

Teams remarks: As is evident from the pair plots, ram is going to be a significant variable to determine the decision boundaries for the models
#checking to see y counts
y.value_counts()
0 500
1 500
2 500
3 500
Name: price_range, dtype: int64
Phase 4: Modelling Strategy
Given this is a classification problem for multiple classes, several models will be used for the comparitive analysis and the best model will be chosen for this project
Model 1: K Means
Model 2: KNN
Model 3: Decision Tree
Model 4: Random Forest
Model 5: XG Boost
Model 6: SVM Classification
Splitting the dataset into train and test data sets for validation
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42, stratify=y) #stratify -> it can reduce the variability of sample statistics
X_train.shape, X_test.shape, y_train.shape, y_test.shape
((1340, 10), (660, 10), (1340,), (660,))
Scaling the train and test datasets
#feature scaling
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train_fc = sc.fit_transform(X_train)
X_test_fc = sc.transform(X_test)
Model 1: K Means
from sklearn.model_selection import GridSearchCV
from sklearn.cluster import KMeans
# Model building
kmeans_mod = KMeans()
kmeans_mod = KMeans(n_clusters = 4, random_state = 0)
kmeans_mod.fit(X)
kmeans_mod.labels_
array([1, 0, 0, ..., 0, 3, 1])
from datetime import datetime
start_time = datetime.now()
from datetime import timedelta
start = timeit.default_timer()
kmeans_mod = KMeans()
kmeans_mod = KMeans(n_clusters = 4, random_state = 0)
kmeans_mod.fit(X)
kmeans_mod.labels_
kmeans_mod.predict(X)
clusters = kmeans_mod.labels_
Kmeansscore = silhouette_score(X, clusters, metric='euclidean')
Kmeansscore
stop = timeit.default_timer()
execution_time = stop - start
print("Program Executed in "+str(execution_time), 'seconds') # It returns time in seconds
#print(timedelta(seconds=end_time - start_time))
#print('Duration: {}'.format(end_time - start_time))
Program Executed in 0.35800029999995786 seconds
start = timeit.default_timer()
kmeans_mod = KMeans()
kmeans_mod = KMeans(n_clusters = 4, random_state = 0)
kmeans_mod.fit(X)
kmeans_mod.labels_
kmeans_mod.predict(X)
clusters = kmeans_mod.labels_
Kmeansscore = silhouette_score(X, clusters, metric='euclidean')
Kmeansscore
stop = timeit.default_timer()
execution_time = stop - start
print("Program Executed in "+str(round(execution_time, 2)), 'seconds') # It returns time in seconds
#print(timedelta(seconds=end_time - start_time))
#print('Duration: {}'.format(end_time - start_time))
Program Executed in 0.39 seconds
#scatterplot analysis
sns.scatterplot(x = 'ram', y = 'battery_power', data = X, hue=y, style= kmeans_mod.labels_, palette=['orange', 'blue', 'red', 'green'])
plt.scatter(kmeans_mod.cluster_centers_[:, 0], kmeans_mod.cluster_centers_[:,1], marker='X', s=200, c='red')
plt.savefig('KMeans Scatterplot.png', dpi = 300)
plt.show()
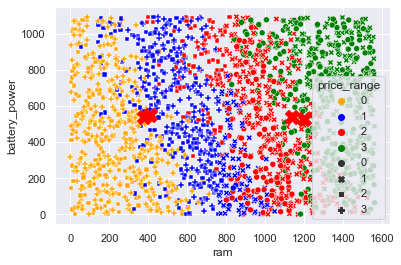
Teams Remarks: Data seems to be overlapping quite a bit at the boundaries (no clear and distinct boundaries) so K Means does not do well in such scenarios as the neighbours are mingled near the boundaries
kmeans_mod.predict(X)
array([1, 0, 0, ..., 0, 3, 1])
#checking the model score
from sklearn.metrics import silhouette_score
clusters = kmeans_mod.labels_
Kmeansscore = silhouette_score(X, clusters, metric='euclidean')
Kmeansscore
0.22549709096550374
Pickling The Model (Save and Load The Model)
import pickle
with open('kmeans_pickle', 'wb') as f:
pickle.dump(kmeans_mod, f)
with open('kmeans_pickle', 'rb') as f:
model_kmeans = pickle.load(f)
model_kmeans.predict(X) #testing the model. reason: to make sure the pickle output showing the same as the current model
array([1, 0, 0, ..., 0, 3, 1])
Teams Remarks: As expected the silhouette score is very low indicating that this modelling technique is not apt for this project __ __
Model 2: KNN
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report
# Model building
knn = KNeighborsClassifier()
# Search parameters
param = range(75, 200, 2)
# Sets up GridSearchCV object and stores it in grid variable
grid = GridSearchCV(knn,{'n_neighbors': param})
# Fits the grid object and gets the best model
best_knn = grid.fit(X_train,y_train).best_estimator_
# Displays the optimum model
best_knn
KNeighborsClassifier(n_neighbors=93)
Grid search found the optimal k value to be 93, using that for the final model below and testing for accuracy
knn = KNeighborsClassifier(n_neighbors=93)
knn.fit(X_train, y_train)
knn_predicted = knn.predict(X_test)
knn_conf_matrix = confusion_matrix(y_test, knn_predicted)
knn_acc_score = accuracy_score(y_test, knn_predicted)
print("confusion matrix")
print(knn_conf_matrix)
print("-------------------------------------------")
print("Accuracy of K-NeighborsClassifier:",knn_acc_score*100,'\n')
print("-------------------------------------------")
print(classification_report(y_test,knn_predicted))
confusion matrix
[[155 10 0 0]
[ 3 154 8 0]
[ 0 11 145 9]
[ 0 0 19 146]]
-------------------------------------------
Accuracy of K-NeighborsClassifier: 90.9090909090909
-------------------------------------------
precision recall f1-score support
0 0.98 0.94 0.96 165
1 0.88 0.93 0.91 165
2 0.84 0.88 0.86 165
3 0.94 0.88 0.91 165
accuracy 0.91 660
macro avg 0.91 0.91 0.91 660
weighted avg 0.91 0.91 0.91 660
# prediction
knn_train_predict = knn.predict(X_train)
knn_test_predict = knn.predict(X_test)
# training and testing accuracy scores
print(accuracy_score(y_train ,knn_train_predict))
KNNscore = accuracy_score(y_test ,knn_test_predict)
KNNscore
0.9097014925373135
0.9090909090909091
Pickling The Model (Save and Load The Model)
with open('knn_pickle', 'wb') as f:
pickle.dump(knn, f)
with open('knn_pickle', 'rb') as f:
model_knn = pickle.load(f)
model_knn.predict(X_test) #testing the model. reason: to make sure the pickle output showing the same as the current model
array([0, 1, 3, 3, 1, 3, 3, 1, 2, 2, 3, 3, 2, 1, 0, 3, 2, 0, 3, 3, 0, 2,
1, 1, 3, 3, 1, 0, 1, 3, 3, 1, 3, 3, 1, 2, 3, 1, 3, 3, 2, 2, 2, 3,
2, 2, 2, 0, 3, 0, 0, 2, 0, 3, 1, 0, 1, 3, 2, 1, 0, 2, 2, 1, 0, 2,
0, 2, 0, 0, 1, 2, 3, 1, 1, 0, 2, 1, 1, 3, 2, 1, 0, 1, 3, 1, 2, 0,
0, 0, 0, 3, 3, 0, 2, 1, 3, 2, 3, 1, 0, 2, 2, 0, 0, 3, 2, 2, 1, 0,
3, 2, 1, 1, 0, 1, 2, 3, 3, 2, 1, 0, 1, 0, 3, 2, 0, 0, 0, 2, 3, 1,
2, 0, 1, 0, 2, 1, 0, 2, 1, 1, 3, 2, 3, 2, 3, 0, 3, 1, 2, 1, 2, 1,
2, 0, 0, 1, 3, 3, 2, 2, 2, 0, 2, 2, 1, 0, 1, 2, 3, 2, 0, 3, 3, 1,
1, 0, 1, 2, 1, 0, 2, 2, 1, 3, 2, 0, 2, 3, 3, 0, 1, 2, 2, 2, 3, 1,
3, 0, 2, 2, 0, 3, 1, 1, 1, 3, 1, 2, 2, 1, 3, 2, 2, 3, 3, 2, 2, 0,
1, 0, 3, 3, 3, 2, 0, 1, 3, 1, 3, 0, 3, 2, 0, 0, 2, 1, 0, 2, 0, 2,
0, 1, 3, 3, 2, 0, 1, 1, 0, 0, 3, 0, 3, 2, 3, 1, 1, 1, 1, 1, 3, 1,
0, 1, 0, 1, 1, 3, 1, 1, 2, 3, 3, 1, 2, 3, 2, 2, 1, 1, 1, 2, 1, 1,
0, 3, 0, 3, 2, 1, 1, 3, 0, 2, 3, 0, 0, 0, 2, 1, 1, 2, 2, 3, 1, 1,
1, 1, 0, 0, 1, 1, 2, 2, 2, 0, 2, 0, 2, 3, 3, 0, 2, 3, 0, 2, 3, 2,
1, 0, 1, 2, 2, 0, 1, 0, 1, 0, 0, 2, 3, 3, 1, 2, 2, 0, 0, 2, 3, 1,
2, 2, 3, 2, 2, 3, 1, 0, 0, 3, 0, 3, 0, 0, 0, 3, 1, 3, 3, 2, 1, 1,
1, 3, 0, 1, 1, 1, 1, 3, 0, 1, 1, 2, 0, 0, 2, 0, 2, 1, 2, 3, 2, 3,
1, 2, 3, 0, 3, 3, 0, 3, 0, 0, 0, 3, 0, 0, 1, 0, 1, 0, 2, 2, 1, 0,
2, 3, 1, 2, 3, 2, 3, 1, 3, 3, 2, 2, 2, 3, 1, 3, 2, 0, 3, 2, 0, 1,
0, 2, 3, 0, 0, 0, 1, 0, 0, 1, 2, 1, 0, 1, 1, 2, 1, 2, 1, 1, 0, 1,
3, 3, 0, 0, 1, 3, 2, 3, 3, 1, 1, 3, 1, 2, 1, 3, 2, 0, 2, 0, 1, 1,
1, 3, 1, 3, 3, 1, 3, 2, 3, 3, 2, 1, 3, 2, 2, 3, 0, 1, 1, 0, 0, 2,
2, 1, 1, 1, 3, 0, 0, 2, 3, 2, 2, 1, 1, 0, 1, 1, 0, 0, 1, 3, 2, 2,
3, 1, 0, 1, 0, 0, 0, 0, 3, 1, 3, 3, 1, 0, 0, 1, 2, 2, 3, 0, 2, 1,
2, 3, 2, 0, 1, 3, 2, 2, 3, 3, 3, 1, 3, 3, 2, 2, 1, 2, 0, 0, 0, 1,
0, 3, 2, 0, 0, 2, 1, 0, 1, 0, 1, 3, 3, 0, 2, 3, 1, 0, 1, 3, 1, 2,
2, 2, 0, 2, 3, 3, 3, 2, 2, 2, 2, 0, 2, 1, 3, 2, 1, 3, 2, 0, 2, 2,
2, 3, 1, 1, 0, 3, 1, 3, 2, 1, 0, 1, 1, 2, 3, 0, 3, 0, 2, 0, 0, 0,
1, 0, 1, 2, 3, 1, 0, 3, 0, 2, 2, 2, 2, 0, 2, 1, 1, 3, 2, 1, 1, 0],
dtype=int64)
Model 3: Decision Tree
# Model building
from sklearn.tree import DecisionTreeClassifier
Deci_Tree_model = DecisionTreeClassifier()
parameters = {'max_depth':[3,5,10,20,30],
'random_state': [0,1,2,3,4]
}
grid = GridSearchCV(Deci_Tree_model,parameters,cv=5,verbose=1)
grid.fit(X_train,y_train)
Fitting 5 folds for each of 25 candidates, totalling 125 fits
GridSearchCV(cv=5, estimator=DecisionTreeClassifier(),
param_grid={'max_depth': [3, 5, 10, 20, 30],
'random_state': [0, 1, 2, 3, 4]},
verbose=1)
# printing best parameters from the above grid analysis
print(grid.best_params_)
{'max_depth': 10, 'random_state': 3}
Grid search has provided the best parameters for the tree, so using these parameters for the final tree below, the depth of 10 seems to be ok given there are 10 predictor variables
# Model building
Deci_Tree_best_model = DecisionTreeClassifier(max_depth=10, random_state=3)
# Model fitting to the datasets
Deci_Tree_best_model.fit(X_train , y_train)
DecisionTreeClassifier(max_depth=10, random_state=3)
# Training and testing of the model
train_predict = Deci_Tree_best_model.predict(X_train)
test_predict = Deci_Tree_best_model.predict(X_test)
# checking the accuracy scores of the model
Deci_Tree_train_accuracyscore = accuracy_score(y_train ,train_predict)
Deci_Tree_test_accuracyscore = accuracy_score(y_test, test_predict)
print(Deci_Tree_train_accuracyscore, Deci_Tree_test_accuracyscore)
0.994776119402985 0.8181818181818182
Pickling The Model (Save and Load The Model)
with open('dec_tree_pickle', 'wb') as f:
pickle.dump(Deci_Tree_best_model, f)
with open('dec_tree_pickle', 'rb') as f:
model_dectree = pickle.load(f)
model_dectree.predict(X_test) #testing the model. reason: to make sure the pickle output showing the same as the current model
array([0, 2, 3, 2, 1, 3, 3, 1, 2, 2, 3, 3, 1, 1, 0, 3, 2, 0, 3, 3, 0, 2,
1, 1, 3, 3, 2, 1, 1, 3, 3, 2, 3, 3, 1, 1, 3, 1, 2, 3, 2, 2, 2, 3,
1, 3, 2, 0, 3, 1, 0, 2, 0, 3, 2, 0, 2, 3, 2, 1, 1, 2, 2, 0, 0, 2,
1, 3, 0, 0, 0, 2, 3, 1, 2, 0, 2, 1, 1, 3, 3, 1, 1, 2, 3, 0, 2, 0,
0, 0, 0, 3, 3, 0, 3, 1, 3, 2, 3, 1, 0, 3, 2, 0, 0, 3, 3, 2, 1, 0,
2, 1, 1, 1, 1, 0, 2, 3, 3, 2, 0, 0, 1, 0, 3, 1, 1, 0, 0, 2, 3, 2,
2, 1, 1, 0, 2, 1, 0, 2, 0, 0, 3, 2, 3, 2, 3, 0, 3, 1, 2, 1, 3, 1,
2, 1, 0, 0, 3, 2, 2, 2, 2, 0, 2, 2, 1, 0, 1, 2, 3, 2, 0, 3, 3, 2,
1, 0, 1, 2, 1, 1, 2, 3, 0, 3, 3, 0, 2, 2, 3, 0, 1, 2, 2, 2, 3, 2,
3, 0, 2, 2, 0, 3, 1, 1, 1, 3, 1, 2, 1, 1, 3, 2, 2, 3, 3, 1, 3, 0,
1, 0, 3, 3, 3, 2, 0, 2, 2, 2, 3, 0, 3, 2, 0, 0, 2, 0, 1, 3, 0, 2,
0, 2, 3, 3, 1, 0, 1, 1, 1, 0, 3, 0, 3, 2, 3, 1, 2, 1, 1, 1, 3, 1,
0, 0, 0, 1, 1, 3, 2, 1, 2, 3, 3, 1, 2, 3, 2, 1, 1, 1, 1, 1, 1, 1,
1, 3, 0, 3, 3, 1, 1, 3, 0, 2, 3, 0, 0, 0, 2, 1, 1, 2, 3, 3, 1, 0,
1, 1, 0, 0, 0, 1, 3, 2, 1, 0, 2, 1, 2, 3, 2, 0, 3, 3, 0, 2, 2, 1,
1, 0, 1, 2, 3, 0, 1, 0, 1, 0, 0, 2, 3, 3, 1, 2, 2, 0, 1, 2, 3, 1,
2, 2, 3, 2, 3, 3, 1, 0, 0, 3, 0, 3, 0, 0, 0, 3, 1, 3, 3, 2, 2, 1,
1, 3, 0, 1, 0, 1, 1, 3, 0, 1, 1, 3, 0, 0, 2, 0, 1, 1, 2, 3, 2, 3,
1, 2, 3, 0, 3, 3, 0, 2, 0, 0, 0, 3, 0, 0, 0, 0, 2, 0, 3, 2, 0, 0,
2, 3, 2, 2, 3, 3, 3, 1, 2, 3, 2, 2, 2, 3, 0, 3, 3, 0, 3, 1, 0, 1,
0, 3, 3, 0, 0, 0, 1, 0, 0, 0, 2, 1, 0, 2, 1, 2, 1, 2, 1, 1, 0, 1,
3, 2, 0, 0, 1, 3, 2, 3, 2, 1, 0, 2, 0, 3, 1, 3, 2, 0, 2, 0, 1, 1,
1, 3, 1, 3, 3, 1, 3, 2, 3, 3, 2, 1, 3, 2, 2, 3, 0, 1, 1, 0, 0, 2,
2, 1, 1, 1, 3, 1, 0, 2, 3, 2, 2, 1, 2, 0, 1, 2, 0, 0, 1, 3, 2, 2,
3, 2, 0, 1, 0, 0, 0, 0, 3, 1, 3, 3, 2, 0, 0, 0, 2, 2, 3, 0, 1, 1,
2, 2, 2, 0, 1, 3, 2, 2, 2, 3, 3, 1, 3, 2, 3, 2, 1, 2, 0, 0, 0, 0,
0, 3, 2, 0, 0, 2, 1, 0, 1, 0, 1, 3, 3, 0, 3, 3, 1, 0, 1, 3, 1, 2,
2, 2, 0, 2, 2, 3, 2, 2, 1, 3, 1, 1, 2, 1, 2, 2, 1, 3, 3, 0, 2, 2,
2, 2, 2, 1, 0, 3, 1, 3, 2, 1, 0, 2, 1, 3, 3, 0, 2, 0, 2, 0, 0, 0,
1, 0, 1, 2, 3, 2, 0, 3, 0, 2, 2, 2, 2, 0, 2, 1, 1, 3, 2, 0, 1, 0],
dtype=int64)
The accuracy score for test data seems to be lower than for KNN model
# visualising the tree
feature_label = list(X.columns)
target_label = [str(s) for s in y.unique()]
Visualizing the decision tree
from sklearn import tree
fig = plt.figure(figsize=(25,20))
_ = tree.plot_tree(Deci_Tree_best_model,
feature_names=feature_label,
class_names=target_label,
filled=True)
plt.savefig('Decision Tree Visualization.png', dpi = 300)
plt.show()
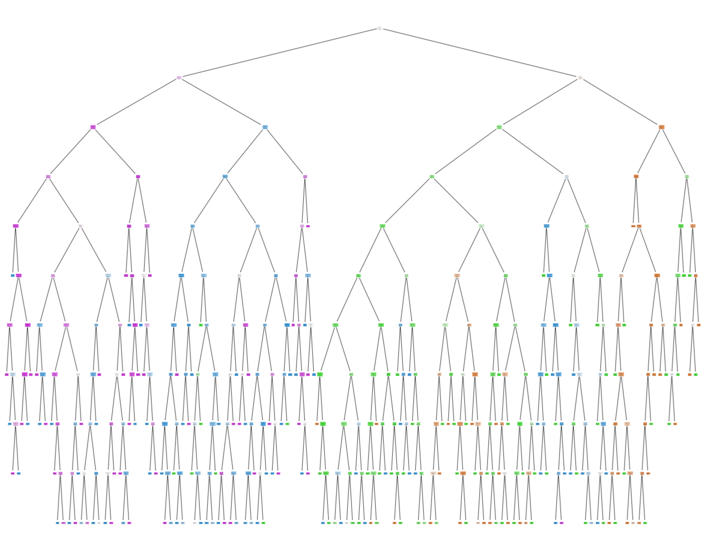
Visualizing the decision boundary.
As the no. of dimensions is 10, reducing the no of dimensions to 2 using PCA to visualize a 2D plot of the boundaries
# visualizing the decision regions
from mlxtend.plotting import plot_decision_regions
from sklearn.decomposition import PCA
a = X_train.to_numpy() #changing df to numpy
b = y_train.to_numpy()
pca = PCA(n_components = 2)
a2 = pca.fit_transform(a)
Deci_Tree_best_model.fit(a2, b)
plot_decision_regions(a2, b, clf=Deci_Tree_best_model, legend=2)
# Adding axes annotations
plt.xlabel('ram')
plt.ylabel('battery_power')
plt.title('Decision Tree Boundaries')
plt.savefig('Decision Tree Boundaries (Using PCA).png', dpi = 300)
plt.show()
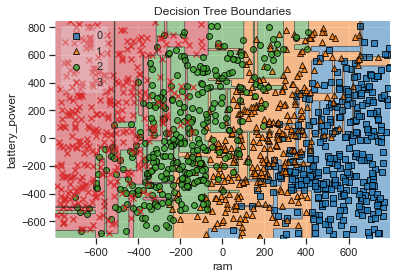
The decision tree model does a better job of demarking the clustered boundaries when compared to the K Means model, hence the improved accuracy score
Model 4: Random Forest
# Model building
from sklearn.ensemble import RandomForestClassifier
RFmodel = RandomForestClassifier() # Hyperparameters tuning
parameters = {'max_depth':[5,10,15,20,30],
'random_state': [0,1,2,3,4],
'n_estimators':[10,30,50,70,100],
'criterion': ['entropy', 'ginni']
}
grid = GridSearchCV(RFmodel,parameters,cv=5,verbose=1)
grid.fit(X_train,y_train)
Fitting 5 folds for each of 250 candidates, totalling 1250 fits
GridSearchCV(cv=5, estimator=RandomForestClassifier(),
param_grid={'criterion': ['entropy', 'ginni'],
'max_depth': [5, 10, 15, 20, 30],
'n_estimators': [10, 30, 50, 70, 100],
'random_state': [0, 1, 2, 3, 4]},
verbose=1)
# Printing the best parameters
print(grid.best_params_)
print(grid.best_estimator_)
{'criterion': 'entropy', 'max_depth': 20, 'n_estimators': 100, 'random_state': 2}
RandomForestClassifier(criterion='entropy', max_depth=20, random_state=2)
# Building the best fit model using the parameters from the grid search
RFBmodel = RandomForestClassifier(random_state=2, max_depth= 20 , n_estimators=50)
RFBmodel.fit(X_train, y_train)
RandomForestClassifier(max_depth=20, n_estimators=50, random_state=2)
# Training and testing the model and checking accuracy scores
RFBtrain_predict = RFBmodel.predict(X_train)
RFBtest_predict = RFBmodel.predict(X_test)
print(accuracy_score(y_train ,RFBtrain_predict))
RFscore = accuracy_score(y_test , RFBtest_predict)
RFscore
1.0
0.896969696969697
# Visualizing the tree
fig = plt.figure(figsize=(25,20))
_ = tree.plot_tree(RFBmodel.estimators_[0],
feature_names=feature_label,
class_names=target_label,
filled=True)
plt.savefig('Random Forest Visualization.png', dpi = 300)
plt.show()
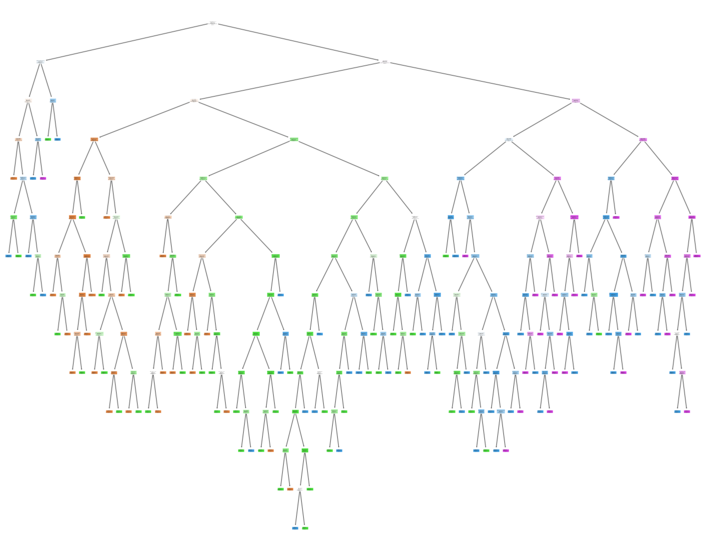
Pickling The Model (Save and Load The Model)
with open('rf_pickle', 'wb') as f:
pickle.dump(RFBmodel, f)
with open('rf_pickle', 'rb') as f:
model_rf = pickle.load(f)
model_rf.predict(X_test) #testing the model. reason: to make sure the pickle output showing the same as the current model
array([0, 2, 3, 2, 1, 3, 3, 1, 2, 2, 3, 3, 2, 1, 0, 3, 2, 0, 3, 3, 0, 2,
2, 1, 3, 3, 1, 0, 1, 3, 3, 1, 3, 3, 1, 1, 3, 1, 3, 3, 2, 2, 2, 3,
1, 2, 2, 0, 3, 0, 1, 2, 0, 3, 2, 0, 1, 3, 2, 1, 0, 2, 2, 1, 0, 2,
0, 2, 0, 0, 1, 1, 3, 1, 2, 0, 2, 1, 1, 3, 2, 1, 1, 2, 3, 1, 2, 0,
0, 0, 0, 3, 3, 0, 3, 1, 3, 2, 3, 1, 0, 2, 2, 0, 0, 3, 3, 2, 1, 0,
2, 1, 1, 0, 0, 0, 2, 3, 3, 2, 0, 0, 1, 0, 3, 2, 0, 0, 0, 2, 3, 1,
2, 0, 1, 0, 2, 1, 0, 2, 1, 1, 3, 2, 2, 2, 3, 0, 3, 1, 2, 1, 3, 1,
2, 0, 0, 0, 3, 3, 2, 2, 2, 0, 2, 2, 1, 0, 1, 2, 3, 2, 0, 3, 3, 2,
1, 0, 1, 2, 1, 0, 2, 2, 1, 3, 3, 0, 2, 3, 3, 0, 1, 1, 2, 1, 3, 2,
3, 0, 2, 2, 0, 3, 1, 1, 1, 3, 2, 2, 2, 1, 3, 2, 2, 3, 3, 1, 2, 0,
1, 0, 3, 3, 3, 2, 0, 1, 3, 1, 3, 0, 3, 2, 0, 0, 2, 1, 1, 2, 0, 2,
0, 1, 3, 3, 2, 0, 1, 1, 0, 0, 3, 0, 3, 2, 3, 0, 1, 1, 1, 1, 3, 1,
0, 1, 0, 1, 1, 3, 1, 1, 2, 3, 3, 1, 2, 2, 2, 2, 1, 1, 1, 2, 1, 2,
0, 3, 0, 3, 2, 1, 1, 3, 0, 2, 3, 0, 0, 0, 1, 2, 1, 2, 2, 3, 1, 1,
1, 1, 0, 0, 1, 1, 2, 2, 1, 0, 2, 0, 2, 3, 3, 0, 2, 3, 0, 2, 2, 2,
1, 0, 1, 2, 2, 0, 1, 0, 1, 0, 0, 2, 3, 3, 1, 2, 2, 0, 0, 2, 3, 1,
2, 2, 3, 2, 3, 3, 1, 0, 0, 3, 0, 3, 0, 0, 0, 3, 1, 3, 3, 2, 2, 1,
1, 3, 0, 1, 0, 1, 1, 3, 0, 1, 1, 3, 0, 0, 2, 0, 1, 1, 2, 3, 2, 3,
2, 2, 3, 0, 3, 3, 0, 3, 0, 0, 0, 3, 0, 0, 1, 0, 1, 0, 2, 2, 1, 0,
2, 3, 2, 2, 3, 2, 3, 1, 3, 3, 1, 2, 2, 3, 1, 3, 3, 0, 3, 2, 0, 1,
0, 2, 3, 0, 0, 0, 1, 0, 0, 0, 3, 1, 0, 1, 1, 2, 0, 3, 1, 1, 0, 2,
3, 3, 0, 0, 1, 3, 2, 3, 2, 1, 1, 3, 0, 3, 1, 3, 2, 0, 2, 0, 1, 1,
1, 3, 1, 3, 3, 1, 3, 2, 3, 3, 2, 1, 3, 2, 2, 3, 0, 1, 1, 0, 0, 2,
2, 1, 1, 1, 3, 1, 0, 2, 3, 2, 2, 1, 1, 0, 1, 1, 0, 0, 1, 3, 2, 2,
3, 1, 0, 1, 0, 0, 0, 0, 3, 1, 3, 3, 1, 0, 0, 0, 3, 2, 3, 0, 2, 1,
2, 3, 2, 0, 1, 3, 2, 2, 3, 3, 3, 1, 3, 2, 2, 2, 1, 2, 0, 0, 0, 0,
0, 3, 2, 0, 0, 2, 1, 0, 0, 0, 1, 3, 3, 0, 2, 3, 1, 0, 1, 3, 1, 2,
2, 2, 0, 2, 3, 3, 2, 2, 1, 3, 2, 0, 2, 1, 3, 2, 1, 3, 2, 0, 2, 2,
2, 3, 1, 1, 0, 3, 1, 3, 2, 1, 0, 2, 1, 2, 3, 0, 3, 0, 3, 0, 0, 0,
1, 1, 1, 2, 2, 1, 0, 3, 0, 2, 2, 3, 2, 0, 2, 1, 1, 3, 2, 0, 1, 0],
dtype=int64)
Model 5: XG Boost
#! pip install xgboost
# import XGBoost
import xgboost as xgb
# Model building and using gridsearch to get the optimal parameters
from xgboost import XGBClassifier
params = {
'objective':'binary:logistic',
'max_depth': 4,
'learning_rate': 1.0,
'n_estimators':100
}
# instantiate the classifier
xgb_clf = XGBClassifier(**params)
# fit the classifier to the training data
xgb_clf.fit(X_train, y_train)
[13:42:50] WARNING: C:/Users/Administrator/workspace/xgboost-win64_release_1.4.0/src/learner.cc:1095: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'multi:softprob' was changed from 'merror' to 'mlogloss'. Explicitly set eval_metric if you'd like to restore the old behavior.
XGBClassifier(base_score=0.5, booster='gbtree', colsample_bylevel=1,
colsample_bynode=1, colsample_bytree=1, gamma=0, gpu_id=-1,
importance_type='gain', interaction_constraints='',
learning_rate=1.0, max_delta_step=0, max_depth=4,
min_child_weight=1, missing=nan, monotone_constraints='()',
n_estimators=100, n_jobs=4, num_parallel_tree=1,
objective='multi:softprob', random_state=0, reg_alpha=0,
reg_lambda=1, scale_pos_weight=None, subsample=1,
tree_method='exact', validate_parameters=1, verbosity=None)
# Prediction using the best fit model and checking accuracy scores
y_pred = xgb_clf.predict(X_test)
XGBscore = accuracy_score(y_test, y_pred)
XGBscore
0.9303030303030303
Pickling The Model (Save and Load The Model)
with open('xgb_pickle', 'wb') as f:
pickle.dump(xgb_clf, f)
with open('xgb_pickle', 'rb') as f:
model_xgb = pickle.load(f)
model_xgb.predict(X_test) #testing the model. reason: to make sure the pickle output showing the same as the current model
array([0, 2, 3, 2, 1, 3, 3, 1, 2, 2, 3, 3, 2, 1, 0, 3, 2, 0, 2, 3, 0, 2,
2, 1, 3, 3, 1, 0, 1, 3, 3, 1, 3, 3, 1, 1, 3, 1, 3, 3, 2, 2, 2, 3,
2, 2, 2, 0, 3, 0, 0, 2, 0, 3, 2, 0, 1, 3, 2, 0, 0, 2, 2, 1, 0, 2,
0, 2, 0, 0, 1, 1, 3, 1, 1, 0, 2, 1, 2, 3, 2, 1, 1, 2, 3, 1, 2, 0,
0, 0, 0, 3, 3, 0, 3, 1, 3, 2, 3, 1, 0, 2, 2, 0, 0, 3, 2, 2, 1, 0,
2, 1, 1, 1, 0, 0, 2, 3, 3, 2, 0, 0, 1, 0, 3, 2, 0, 0, 0, 2, 3, 1,
2, 0, 1, 0, 2, 1, 0, 2, 1, 1, 3, 2, 3, 2, 3, 0, 3, 1, 2, 1, 2, 1,
2, 0, 0, 1, 3, 3, 2, 2, 2, 0, 2, 2, 1, 0, 1, 2, 3, 2, 0, 3, 3, 2,
1, 0, 1, 2, 1, 0, 2, 2, 0, 3, 2, 0, 2, 3, 3, 0, 1, 2, 2, 2, 3, 1,
3, 0, 2, 2, 0, 3, 1, 1, 1, 3, 1, 2, 2, 1, 3, 2, 2, 3, 3, 1, 2, 0,
1, 0, 3, 3, 3, 2, 0, 1, 3, 1, 3, 0, 3, 2, 0, 0, 2, 1, 0, 2, 0, 2,
0, 1, 3, 3, 1, 0, 2, 1, 0, 0, 3, 0, 3, 2, 3, 1, 2, 1, 1, 1, 3, 1,
0, 1, 0, 1, 1, 3, 1, 1, 2, 3, 3, 1, 3, 3, 2, 1, 2, 1, 1, 2, 1, 2,
0, 3, 0, 3, 3, 1, 1, 3, 0, 2, 3, 0, 0, 0, 2, 1, 1, 2, 3, 2, 1, 1,
1, 1, 0, 0, 1, 1, 2, 2, 1, 0, 2, 0, 2, 3, 3, 0, 2, 3, 0, 2, 2, 2,
1, 0, 1, 2, 2, 0, 1, 0, 1, 0, 0, 2, 3, 3, 1, 2, 2, 0, 0, 1, 3, 1,
2, 2, 3, 2, 3, 3, 1, 0, 0, 3, 0, 3, 0, 0, 0, 3, 1, 3, 3, 2, 1, 1,
1, 3, 0, 1, 0, 1, 1, 3, 0, 1, 1, 3, 0, 0, 2, 0, 2, 1, 2, 3, 2, 3,
1, 2, 3, 0, 3, 3, 0, 3, 0, 0, 0, 3, 0, 0, 1, 0, 1, 0, 2, 2, 1, 0,
3, 3, 2, 2, 3, 2, 3, 1, 3, 3, 1, 2, 2, 3, 1, 3, 3, 0, 3, 1, 0, 1,
0, 2, 3, 0, 0, 0, 1, 1, 0, 0, 3, 1, 0, 1, 1, 3, 1, 3, 1, 1, 0, 1,
3, 3, 0, 0, 1, 3, 2, 3, 3, 1, 1, 2, 0, 3, 1, 3, 2, 0, 2, 0, 1, 1,
2, 3, 1, 3, 3, 1, 3, 2, 3, 3, 2, 1, 3, 2, 2, 3, 0, 1, 1, 0, 0, 2,
2, 1, 1, 1, 3, 0, 0, 2, 3, 2, 2, 1, 1, 0, 1, 1, 0, 0, 1, 3, 2, 2,
3, 1, 0, 1, 0, 0, 0, 0, 3, 1, 3, 3, 2, 0, 0, 1, 3, 2, 3, 0, 2, 1,
2, 3, 2, 0, 1, 3, 2, 2, 3, 3, 3, 1, 3, 2, 2, 2, 1, 2, 0, 0, 0, 1,
0, 3, 2, 0, 0, 2, 1, 0, 0, 0, 1, 3, 3, 0, 2, 3, 1, 0, 1, 3, 1, 2,
2, 2, 0, 3, 3, 3, 2, 2, 2, 2, 2, 0, 2, 1, 3, 2, 1, 3, 2, 0, 2, 2,
2, 3, 1, 1, 0, 3, 1, 3, 2, 1, 0, 2, 1, 2, 3, 0, 3, 0, 3, 0, 0, 0,
1, 0, 1, 2, 2, 1, 0, 3, 0, 2, 2, 3, 2, 0, 2, 1, 1, 3, 2, 0, 1, 0],
dtype=int64)
Model 6: SVM Classification
from sklearn.svm import SVC
from sklearn.metrics import classification_report
from mlxtend.plotting import plot_decision_regions
from sklearn.svm import SVC
# Building the best fit model using the gridsearch methodology
param_grid = {'C': [0.1, 1, 10, 100, 1000],
'gamma': [1, 0.1, 0.01, 0.001, 0.0001],
'kernel': ['rbf', 'poly']}
grid = GridSearchCV(SVC(), param_grid, refit = True, verbose = 3)
# fitting the model for grid search
grid.fit(X_train, y_train)
Fitting 5 folds for each of 50 candidates, totalling 250 fits
[CV 1/5] END ........C=0.1, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 2/5] END ........C=0.1, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 3/5] END ........C=0.1, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 4/5] END ........C=0.1, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 5/5] END ........C=0.1, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 1/5] END .......C=0.1, gamma=1, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END .......C=0.1, gamma=1, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END .......C=0.1, gamma=1, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END .......C=0.1, gamma=1, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END .......C=0.1, gamma=1, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ......C=0.1, gamma=0.1, kernel=rbf;, score=0.313 total time= 0.2s
[CV 2/5] END ......C=0.1, gamma=0.1, kernel=rbf;, score=0.306 total time= 0.2s
[CV 3/5] END ......C=0.1, gamma=0.1, kernel=rbf;, score=0.313 total time= 0.2s
[CV 4/5] END ......C=0.1, gamma=0.1, kernel=rbf;, score=0.299 total time= 0.2s
[CV 5/5] END ......C=0.1, gamma=0.1, kernel=rbf;, score=0.295 total time= 0.2s
[CV 1/5] END .....C=0.1, gamma=0.1, kernel=poly;, score=0.970 total time= 0.1s
[CV 2/5] END .....C=0.1, gamma=0.1, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END .....C=0.1, gamma=0.1, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END .....C=0.1, gamma=0.1, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END .....C=0.1, gamma=0.1, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END .....C=0.1, gamma=0.01, kernel=rbf;, score=0.500 total time= 0.2s
[CV 2/5] END .....C=0.1, gamma=0.01, kernel=rbf;, score=0.500 total time= 0.2s
[CV 3/5] END .....C=0.1, gamma=0.01, kernel=rbf;, score=0.496 total time= 0.2s
[CV 4/5] END .....C=0.1, gamma=0.01, kernel=rbf;, score=0.496 total time= 0.2s
[CV 5/5] END .....C=0.1, gamma=0.01, kernel=rbf;, score=0.683 total time= 0.2s
[CV 1/5] END ....C=0.1, gamma=0.01, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END ....C=0.1, gamma=0.01, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ....C=0.1, gamma=0.01, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ....C=0.1, gamma=0.01, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ....C=0.1, gamma=0.01, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ....C=0.1, gamma=0.001, kernel=rbf;, score=0.265 total time= 0.1s
[CV 2/5] END ....C=0.1, gamma=0.001, kernel=rbf;, score=0.257 total time= 0.1s
[CV 3/5] END ....C=0.1, gamma=0.001, kernel=rbf;, score=0.261 total time= 0.2s
[CV 4/5] END ....C=0.1, gamma=0.001, kernel=rbf;, score=0.265 total time= 0.1s
[CV 5/5] END ....C=0.1, gamma=0.001, kernel=rbf;, score=0.272 total time= 0.1s
[CV 1/5] END ...C=0.1, gamma=0.001, kernel=poly;, score=0.970 total time= 0.1s
[CV 2/5] END ...C=0.1, gamma=0.001, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ...C=0.1, gamma=0.001, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ...C=0.1, gamma=0.001, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ...C=0.1, gamma=0.001, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ...C=0.1, gamma=0.0001, kernel=rbf;, score=0.698 total time= 0.1s
[CV 2/5] END ...C=0.1, gamma=0.0001, kernel=rbf;, score=0.646 total time= 0.1s
[CV 3/5] END ...C=0.1, gamma=0.0001, kernel=rbf;, score=0.694 total time= 0.1s
[CV 4/5] END ...C=0.1, gamma=0.0001, kernel=rbf;, score=0.552 total time= 0.1s
[CV 5/5] END ...C=0.1, gamma=0.0001, kernel=rbf;, score=0.754 total time= 0.1s
[CV 1/5] END ..C=0.1, gamma=0.0001, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END ..C=0.1, gamma=0.0001, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ..C=0.1, gamma=0.0001, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ..C=0.1, gamma=0.0001, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ..C=0.1, gamma=0.0001, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ..........C=1, gamma=1, kernel=rbf;, score=0.250 total time= 0.1s
[CV 2/5] END ..........C=1, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 3/5] END ..........C=1, gamma=1, kernel=rbf;, score=0.250 total time= 0.1s
[CV 4/5] END ..........C=1, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 5/5] END ..........C=1, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 1/5] END .........C=1, gamma=1, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END .........C=1, gamma=1, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END .........C=1, gamma=1, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END .........C=1, gamma=1, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END .........C=1, gamma=1, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ........C=1, gamma=0.1, kernel=rbf;, score=0.295 total time= 0.2s
[CV 2/5] END ........C=1, gamma=0.1, kernel=rbf;, score=0.284 total time= 0.2s
[CV 3/5] END ........C=1, gamma=0.1, kernel=rbf;, score=0.295 total time= 0.1s
[CV 4/5] END ........C=1, gamma=0.1, kernel=rbf;, score=0.291 total time= 0.2s
[CV 5/5] END ........C=1, gamma=0.1, kernel=rbf;, score=0.295 total time= 0.1s
[CV 1/5] END .......C=1, gamma=0.1, kernel=poly;, score=0.970 total time= 0.1s
[CV 2/5] END .......C=1, gamma=0.1, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END .......C=1, gamma=0.1, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END .......C=1, gamma=0.1, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END .......C=1, gamma=0.1, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END .......C=1, gamma=0.01, kernel=rbf;, score=0.250 total time= 0.2s
[CV 2/5] END .......C=1, gamma=0.01, kernel=rbf;, score=0.254 total time= 0.2s
[CV 3/5] END .......C=1, gamma=0.01, kernel=rbf;, score=0.250 total time= 0.2s
[CV 4/5] END .......C=1, gamma=0.01, kernel=rbf;, score=0.250 total time= 0.2s
[CV 5/5] END .......C=1, gamma=0.01, kernel=rbf;, score=0.254 total time= 0.2s
[CV 1/5] END ......C=1, gamma=0.01, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END ......C=1, gamma=0.01, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ......C=1, gamma=0.01, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ......C=1, gamma=0.01, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ......C=1, gamma=0.01, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ......C=1, gamma=0.001, kernel=rbf;, score=0.313 total time= 0.2s
[CV 2/5] END ......C=1, gamma=0.001, kernel=rbf;, score=0.310 total time= 0.2s
[CV 3/5] END ......C=1, gamma=0.001, kernel=rbf;, score=0.310 total time= 0.1s
[CV 4/5] END ......C=1, gamma=0.001, kernel=rbf;, score=0.332 total time= 0.2s
[CV 5/5] END ......C=1, gamma=0.001, kernel=rbf;, score=0.306 total time= 0.1s
[CV 1/5] END .....C=1, gamma=0.001, kernel=poly;, score=0.970 total time= 0.1s
[CV 2/5] END .....C=1, gamma=0.001, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END .....C=1, gamma=0.001, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END .....C=1, gamma=0.001, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END .....C=1, gamma=0.001, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END .....C=1, gamma=0.0001, kernel=rbf;, score=0.899 total time= 0.1s
[CV 2/5] END .....C=1, gamma=0.0001, kernel=rbf;, score=0.896 total time= 0.1s
[CV 3/5] END .....C=1, gamma=0.0001, kernel=rbf;, score=0.892 total time= 0.1s
[CV 4/5] END .....C=1, gamma=0.0001, kernel=rbf;, score=0.825 total time= 0.2s
[CV 5/5] END .....C=1, gamma=0.0001, kernel=rbf;, score=0.858 total time= 0.1s
[CV 1/5] END ....C=1, gamma=0.0001, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END ....C=1, gamma=0.0001, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ....C=1, gamma=0.0001, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ....C=1, gamma=0.0001, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ....C=1, gamma=0.0001, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END .........C=10, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 2/5] END .........C=10, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 3/5] END .........C=10, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 4/5] END .........C=10, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 5/5] END .........C=10, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 1/5] END ........C=10, gamma=1, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END ........C=10, gamma=1, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ........C=10, gamma=1, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ........C=10, gamma=1, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ........C=10, gamma=1, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END .......C=10, gamma=0.1, kernel=rbf;, score=0.295 total time= 0.2s
[CV 2/5] END .......C=10, gamma=0.1, kernel=rbf;, score=0.284 total time= 0.2s
[CV 3/5] END .......C=10, gamma=0.1, kernel=rbf;, score=0.295 total time= 0.2s
[CV 4/5] END .......C=10, gamma=0.1, kernel=rbf;, score=0.291 total time= 0.2s
[CV 5/5] END .......C=10, gamma=0.1, kernel=rbf;, score=0.295 total time= 0.2s
[CV 1/5] END ......C=10, gamma=0.1, kernel=poly;, score=0.970 total time= 0.1s
[CV 2/5] END ......C=10, gamma=0.1, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ......C=10, gamma=0.1, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ......C=10, gamma=0.1, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ......C=10, gamma=0.1, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ......C=10, gamma=0.01, kernel=rbf;, score=0.250 total time= 0.2s
[CV 2/5] END ......C=10, gamma=0.01, kernel=rbf;, score=0.254 total time= 0.2s
[CV 3/5] END ......C=10, gamma=0.01, kernel=rbf;, score=0.250 total time= 0.2s
[CV 4/5] END ......C=10, gamma=0.01, kernel=rbf;, score=0.250 total time= 0.1s
[CV 5/5] END ......C=10, gamma=0.01, kernel=rbf;, score=0.254 total time= 0.1s
[CV 1/5] END .....C=10, gamma=0.01, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END .....C=10, gamma=0.01, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END .....C=10, gamma=0.01, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END .....C=10, gamma=0.01, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END .....C=10, gamma=0.01, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END .....C=10, gamma=0.001, kernel=rbf;, score=0.317 total time= 0.2s
[CV 2/5] END .....C=10, gamma=0.001, kernel=rbf;, score=0.317 total time= 0.2s
[CV 3/5] END .....C=10, gamma=0.001, kernel=rbf;, score=0.321 total time= 0.1s
[CV 4/5] END .....C=10, gamma=0.001, kernel=rbf;, score=0.343 total time= 0.1s
[CV 5/5] END .....C=10, gamma=0.001, kernel=rbf;, score=0.317 total time= 0.1s
[CV 1/5] END ....C=10, gamma=0.001, kernel=poly;, score=0.970 total time= 0.1s
[CV 2/5] END ....C=10, gamma=0.001, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ....C=10, gamma=0.001, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ....C=10, gamma=0.001, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ....C=10, gamma=0.001, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ....C=10, gamma=0.0001, kernel=rbf;, score=0.899 total time= 0.1s
[CV 2/5] END ....C=10, gamma=0.0001, kernel=rbf;, score=0.899 total time= 0.1s
[CV 3/5] END ....C=10, gamma=0.0001, kernel=rbf;, score=0.896 total time= 0.1s
[CV 4/5] END ....C=10, gamma=0.0001, kernel=rbf;, score=0.828 total time= 0.1s
[CV 5/5] END ....C=10, gamma=0.0001, kernel=rbf;, score=0.862 total time= 0.1s
[CV 1/5] END ...C=10, gamma=0.0001, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END ...C=10, gamma=0.0001, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ...C=10, gamma=0.0001, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ...C=10, gamma=0.0001, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ...C=10, gamma=0.0001, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ........C=100, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 2/5] END ........C=100, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 3/5] END ........C=100, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 4/5] END ........C=100, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 5/5] END ........C=100, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 1/5] END .......C=100, gamma=1, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END .......C=100, gamma=1, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END .......C=100, gamma=1, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END .......C=100, gamma=1, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END .......C=100, gamma=1, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ......C=100, gamma=0.1, kernel=rbf;, score=0.295 total time= 0.2s
[CV 2/5] END ......C=100, gamma=0.1, kernel=rbf;, score=0.284 total time= 0.2s
[CV 3/5] END ......C=100, gamma=0.1, kernel=rbf;, score=0.295 total time= 0.1s
[CV 4/5] END ......C=100, gamma=0.1, kernel=rbf;, score=0.291 total time= 0.2s
[CV 5/5] END ......C=100, gamma=0.1, kernel=rbf;, score=0.295 total time= 0.2s
[CV 1/5] END .....C=100, gamma=0.1, kernel=poly;, score=0.970 total time= 0.1s
[CV 2/5] END .....C=100, gamma=0.1, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END .....C=100, gamma=0.1, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END .....C=100, gamma=0.1, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END .....C=100, gamma=0.1, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END .....C=100, gamma=0.01, kernel=rbf;, score=0.250 total time= 0.2s
[CV 2/5] END .....C=100, gamma=0.01, kernel=rbf;, score=0.254 total time= 0.1s
[CV 3/5] END .....C=100, gamma=0.01, kernel=rbf;, score=0.250 total time= 0.2s
[CV 4/5] END .....C=100, gamma=0.01, kernel=rbf;, score=0.250 total time= 0.2s
[CV 5/5] END .....C=100, gamma=0.01, kernel=rbf;, score=0.254 total time= 0.1s
[CV 1/5] END ....C=100, gamma=0.01, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END ....C=100, gamma=0.01, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ....C=100, gamma=0.01, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ....C=100, gamma=0.01, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ....C=100, gamma=0.01, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ....C=100, gamma=0.001, kernel=rbf;, score=0.317 total time= 0.2s
[CV 2/5] END ....C=100, gamma=0.001, kernel=rbf;, score=0.317 total time= 0.1s
[CV 3/5] END ....C=100, gamma=0.001, kernel=rbf;, score=0.321 total time= 0.2s
[CV 4/5] END ....C=100, gamma=0.001, kernel=rbf;, score=0.343 total time= 0.2s
[CV 5/5] END ....C=100, gamma=0.001, kernel=rbf;, score=0.317 total time= 0.1s
[CV 1/5] END ...C=100, gamma=0.001, kernel=poly;, score=0.970 total time= 0.1s
[CV 2/5] END ...C=100, gamma=0.001, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ...C=100, gamma=0.001, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ...C=100, gamma=0.001, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ...C=100, gamma=0.001, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ...C=100, gamma=0.0001, kernel=rbf;, score=0.899 total time= 0.1s
[CV 2/5] END ...C=100, gamma=0.0001, kernel=rbf;, score=0.899 total time= 0.1s
[CV 3/5] END ...C=100, gamma=0.0001, kernel=rbf;, score=0.896 total time= 0.1s
[CV 4/5] END ...C=100, gamma=0.0001, kernel=rbf;, score=0.828 total time= 0.1s
[CV 5/5] END ...C=100, gamma=0.0001, kernel=rbf;, score=0.862 total time= 0.1s
[CV 1/5] END ..C=100, gamma=0.0001, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END ..C=100, gamma=0.0001, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ..C=100, gamma=0.0001, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ..C=100, gamma=0.0001, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ..C=100, gamma=0.0001, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END .......C=1000, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 2/5] END .......C=1000, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 3/5] END .......C=1000, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 4/5] END .......C=1000, gamma=1, kernel=rbf;, score=0.250 total time= 0.1s
[CV 5/5] END .......C=1000, gamma=1, kernel=rbf;, score=0.250 total time= 0.2s
[CV 1/5] END ......C=1000, gamma=1, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END ......C=1000, gamma=1, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ......C=1000, gamma=1, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ......C=1000, gamma=1, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ......C=1000, gamma=1, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END .....C=1000, gamma=0.1, kernel=rbf;, score=0.295 total time= 0.2s
[CV 2/5] END .....C=1000, gamma=0.1, kernel=rbf;, score=0.284 total time= 0.2s
[CV 3/5] END .....C=1000, gamma=0.1, kernel=rbf;, score=0.295 total time= 0.2s
[CV 4/5] END .....C=1000, gamma=0.1, kernel=rbf;, score=0.291 total time= 0.2s
[CV 5/5] END .....C=1000, gamma=0.1, kernel=rbf;, score=0.295 total time= 0.2s
[CV 1/5] END ....C=1000, gamma=0.1, kernel=poly;, score=0.970 total time= 0.1s
[CV 2/5] END ....C=1000, gamma=0.1, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ....C=1000, gamma=0.1, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ....C=1000, gamma=0.1, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ....C=1000, gamma=0.1, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ....C=1000, gamma=0.01, kernel=rbf;, score=0.250 total time= 0.2s
[CV 2/5] END ....C=1000, gamma=0.01, kernel=rbf;, score=0.254 total time= 0.2s
[CV 3/5] END ....C=1000, gamma=0.01, kernel=rbf;, score=0.250 total time= 0.2s
[CV 4/5] END ....C=1000, gamma=0.01, kernel=rbf;, score=0.250 total time= 0.2s
[CV 5/5] END ....C=1000, gamma=0.01, kernel=rbf;, score=0.254 total time= 0.2s
[CV 1/5] END ...C=1000, gamma=0.01, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END ...C=1000, gamma=0.01, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ...C=1000, gamma=0.01, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ...C=1000, gamma=0.01, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ...C=1000, gamma=0.01, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ...C=1000, gamma=0.001, kernel=rbf;, score=0.317 total time= 0.1s
[CV 2/5] END ...C=1000, gamma=0.001, kernel=rbf;, score=0.317 total time= 0.2s
[CV 3/5] END ...C=1000, gamma=0.001, kernel=rbf;, score=0.321 total time= 0.2s
[CV 4/5] END ...C=1000, gamma=0.001, kernel=rbf;, score=0.343 total time= 0.1s
[CV 5/5] END ...C=1000, gamma=0.001, kernel=rbf;, score=0.317 total time= 0.2s
[CV 1/5] END ..C=1000, gamma=0.001, kernel=poly;, score=0.970 total time= 0.1s
[CV 2/5] END ..C=1000, gamma=0.001, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END ..C=1000, gamma=0.001, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END ..C=1000, gamma=0.001, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END ..C=1000, gamma=0.001, kernel=poly;, score=0.929 total time= 0.0s
[CV 1/5] END ..C=1000, gamma=0.0001, kernel=rbf;, score=0.899 total time= 0.1s
[CV 2/5] END ..C=1000, gamma=0.0001, kernel=rbf;, score=0.899 total time= 0.1s
[CV 3/5] END ..C=1000, gamma=0.0001, kernel=rbf;, score=0.896 total time= 0.1s
[CV 4/5] END ..C=1000, gamma=0.0001, kernel=rbf;, score=0.828 total time= 0.1s
[CV 5/5] END ..C=1000, gamma=0.0001, kernel=rbf;, score=0.862 total time= 0.1s
[CV 1/5] END .C=1000, gamma=0.0001, kernel=poly;, score=0.970 total time= 0.0s
[CV 2/5] END .C=1000, gamma=0.0001, kernel=poly;, score=0.948 total time= 0.0s
[CV 3/5] END .C=1000, gamma=0.0001, kernel=poly;, score=0.944 total time= 0.0s
[CV 4/5] END .C=1000, gamma=0.0001, kernel=poly;, score=0.944 total time= 0.0s
[CV 5/5] END .C=1000, gamma=0.0001, kernel=poly;, score=0.929 total time= 0.0s
GridSearchCV(estimator=SVC(),
param_grid={'C': [0.1, 1, 10, 100, 1000],
'gamma': [1, 0.1, 0.01, 0.001, 0.0001],
'kernel': ['rbf', 'poly']},
verbose=3)
# print best parameter after tuning
print(grid.best_params_)
# print how our model looks after hyper-parameter tuning
print(grid.best_estimator_)
{'C': 0.1, 'gamma': 1, 'kernel': 'poly'}
SVC(C=0.1, gamma=1, kernel='poly')
# Running the bestfit model and checking for accuracy scores
svc = SVC(C=0.1, gamma=1, kernel='poly')
svc.fit(X_train, y_train)
svc_predicted = svc.predict(X_test)
svc_conf_matrix = confusion_matrix(y_test, svc_predicted)
svc_acc_score = accuracy_score(y_test, svc_predicted)
print("confussion matrix")
print(svc_conf_matrix)
print("-------------------------------------------")
print("Accuracy of Support Vector Classifier:",svc_acc_score*100,'\n')
print("-------------------------------------------")
print(classification_report(y_test,svc_predicted))
confussion matrix
[[162 3 0 0]
[ 2 158 5 0]
[ 0 2 155 8]
[ 0 0 5 160]]
-------------------------------------------
Accuracy of Support Vector Classifier: 96.21212121212122
-------------------------------------------
precision recall f1-score support
0 0.99 0.98 0.98 165
1 0.97 0.96 0.96 165
2 0.94 0.94 0.94 165
3 0.95 0.97 0.96 165
accuracy 0.96 660
macro avg 0.96 0.96 0.96 660
weighted avg 0.96 0.96 0.96 660
Visualizing the classification boundaries for SVC…but since it is taking a long time to process the code has been commented
#m = X_train.to_numpy() #changing df to numpy
#n = y_train.to_numpy()
#plot_decision_regions(m, n,clf=svc, legend =4)
# Adding axes annotations
#plt.xlabel('ram')
#plt.ylabel('battery_power')
#plt.title('SVM Boundaries')
#plt.show()
# Training and testing the best fit model from the gridsearch
svc_train_predict = svc.predict(X_train)
svc_test_predict = svc.predict(X_test)
# checking the accuracy of the best fit model
print(accuracy_score(y_train ,svc_train_predict))
SVCscore = accuracy_score(y_test ,svc_test_predict)
SVCscore
1.0
0.9621212121212122
Pickling The Model (Save and Load The Model)
with open('svc_pickle', 'wb') as f:
pickle.dump(svc, f)
with open('svc_pickle', 'rb') as f:
model_svc = pickle.load(f)
model_svc.predict(X_test) #testing the model. reason: to make sure the pickle output showing the same as the current model
array([0, 2, 3, 3, 1, 3, 3, 1, 2, 2, 3, 3, 2, 1, 0, 3, 2, 0, 3, 3, 0, 2,
2, 1, 3, 3, 1, 0, 1, 3, 3, 1, 3, 3, 1, 1, 3, 1, 3, 3, 2, 2, 2, 3,
2, 3, 2, 0, 3, 0, 0, 2, 0, 2, 1, 0, 1, 3, 2, 0, 0, 2, 2, 0, 0, 2,
0, 2, 0, 0, 1, 2, 3, 1, 1, 0, 2, 1, 1, 3, 3, 1, 1, 1, 3, 1, 2, 0,
0, 0, 0, 3, 3, 0, 3, 1, 3, 2, 3, 1, 0, 2, 2, 0, 0, 3, 2, 2, 1, 0,
2, 2, 1, 1, 0, 0, 2, 3, 3, 2, 1, 0, 1, 0, 3, 2, 0, 0, 0, 2, 3, 1,
2, 0, 1, 0, 2, 1, 0, 2, 1, 1, 3, 2, 3, 2, 3, 0, 3, 1, 2, 1, 3, 1,
2, 0, 0, 1, 3, 3, 2, 2, 2, 0, 2, 2, 1, 0, 1, 2, 3, 2, 0, 3, 3, 2,
2, 0, 1, 2, 1, 0, 2, 2, 0, 3, 2, 0, 2, 3, 3, 0, 2, 2, 2, 2, 3, 1,
3, 0, 2, 2, 0, 3, 1, 1, 1, 3, 1, 2, 2, 1, 3, 2, 3, 3, 3, 1, 2, 0,
1, 0, 3, 3, 3, 2, 0, 1, 3, 1, 3, 0, 3, 2, 0, 0, 2, 1, 0, 3, 0, 2,
0, 1, 3, 3, 2, 0, 2, 1, 0, 0, 3, 0, 3, 2, 3, 1, 1, 1, 1, 1, 3, 1,
0, 1, 0, 1, 1, 3, 1, 1, 2, 3, 3, 1, 2, 3, 2, 1, 1, 1, 1, 1, 1, 1,
0, 3, 0, 3, 3, 1, 2, 3, 0, 2, 3, 0, 0, 0, 2, 1, 1, 2, 3, 2, 1, 1,
1, 1, 0, 0, 1, 1, 2, 2, 1, 0, 2, 0, 2, 3, 3, 0, 3, 3, 0, 2, 2, 2,
1, 0, 1, 2, 2, 0, 1, 0, 1, 0, 0, 2, 3, 3, 1, 2, 2, 0, 0, 2, 3, 1,
2, 2, 3, 2, 2, 3, 1, 0, 0, 3, 0, 3, 0, 0, 0, 3, 1, 3, 3, 2, 1, 1,
1, 3, 0, 1, 1, 1, 1, 3, 0, 1, 1, 3, 0, 0, 2, 0, 2, 1, 2, 3, 2, 3,
1, 2, 3, 0, 3, 3, 0, 3, 0, 0, 0, 3, 0, 0, 1, 0, 1, 0, 2, 2, 1, 0,
2, 3, 2, 2, 3, 2, 3, 1, 3, 3, 1, 2, 2, 3, 1, 3, 3, 0, 3, 2, 0, 1,
0, 2, 3, 0, 1, 0, 1, 0, 0, 0, 3, 1, 0, 1, 1, 3, 0, 3, 1, 1, 0, 1,
3, 3, 0, 1, 1, 3, 2, 3, 3, 1, 1, 2, 0, 3, 1, 3, 2, 0, 2, 0, 1, 1,
2, 3, 0, 3, 3, 1, 3, 3, 3, 3, 2, 1, 3, 2, 2, 3, 0, 1, 1, 0, 0, 2,
2, 1, 1, 1, 3, 0, 0, 3, 3, 2, 2, 1, 2, 0, 1, 1, 0, 0, 1, 3, 2, 2,
3, 1, 0, 1, 0, 0, 0, 0, 3, 1, 3, 3, 1, 0, 0, 1, 3, 2, 3, 0, 2, 1,
2, 3, 2, 0, 1, 3, 2, 2, 3, 3, 3, 1, 3, 2, 2, 2, 1, 2, 0, 0, 0, 1,
0, 3, 2, 0, 0, 2, 1, 0, 0, 0, 1, 3, 3, 0, 2, 3, 1, 0, 1, 3, 1, 2,
2, 2, 0, 3, 3, 3, 2, 3, 2, 3, 2, 0, 2, 1, 3, 2, 1, 3, 2, 0, 2, 2,
2, 2, 2, 1, 0, 3, 1, 3, 2, 1, 0, 2, 1, 2, 3, 0, 3, 0, 2, 0, 0, 0,
1, 0, 1, 2, 2, 1, 0, 3, 0, 2, 2, 3, 2, 0, 2, 1, 1, 3, 2, 1, 1, 0],
dtype=int64)
Phase 5: Summary of the Analysis, Recommendation, Project Risks
Summary of the Analysis
# Summary of the Accuracy scores for test data
model_ev = pd.DataFrame({'Model': ['K-Means','KNN','Decision Tree','Random Forest',
'XG Boost','SVM Classification'], 'Accuracy (%)': [round(Kmeansscore*100, 3),
round(KNNscore*100, 3),round(Deci_Tree_test_accuracyscore*100, 3),round(RFscore*100, 3),round(XGBscore*100, 3),round(SVCscore*100, 3)]})
model_ev
| Model | Accuracy (%) | |
|---|---|---|
| 0 | K-Means | 22.550 |
| 1 | KNN | 90.909 |
| 2 | Decision Tree | 81.818 |
| 3 | Random Forest | 89.697 |
| 4 | XG Boost | 93.030 |
| 5 | SVM Classification | 96.212 |
colors = ['red','green','blue','c','orange','yellow']
plt.figure(figsize=(15,7))
plt.title("Models Performance", size = 20, fontweight='bold')
plt.xlabel("Models", fontweight='bold')
plt.xticks(rotation=0)
plt.ylabel("Accuracy (%)", fontweight='bold')
plt.bar(model_ev['Model'],model_ev['Accuracy (%)'],color = colors)
plt.savefig('Models Performance.png', dpi = 300)
plt.show()
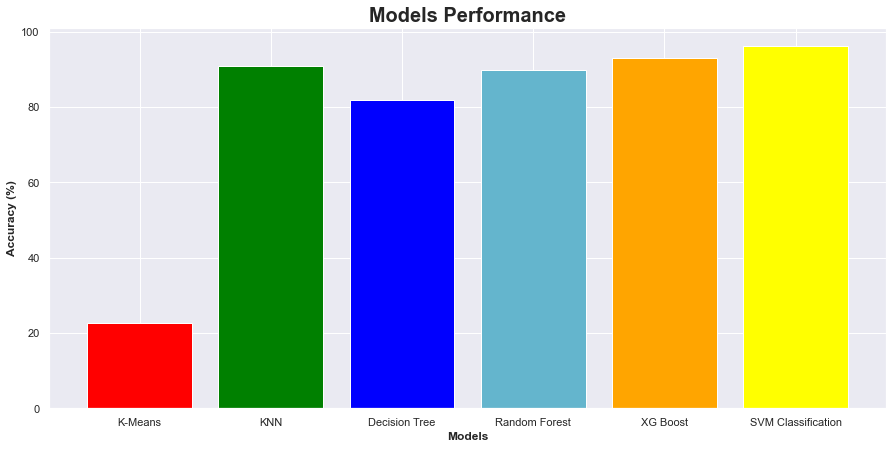
SVM classification has the highest accuracy score
Recommendation
- SVM classification algorithm is the best modeling techinque for this project. This makes sense as the SVM methodology is the best technique for cluttered datasets with more than 3 dimensions. The data seems to be quite overlapping in 2D dimensions near the decision or classification boundaries (as seen in the 2D plots), but the SVM algorithm models the data at higher dimensions where the dataset can be linearly or distinctly divided into unique classes. Hence the higher scores.
- But the disadvantage is that it is very time and resource consuming to run the SVM model. The XG Boost Model gives a comparable accuracy score and takes a lesser time to run the model.
- The model can be further improved by eleminating or combining some of the correlated predictor variables like the (a) screen width and screen height, (b) pixel width and pixel height etc.
- Model can also be simplified and improved by using just the RAM of the cell phones for classification purposes instead of all other variables as RAM alone contributes to almost 92% of the target classification (as per correlation matrix). To limit the scope of the analysis we have chosen not to implement Recommendations 3 & 4.
Project Risks
- We have taken the 10 best features from the feature selection algorithm to reduce the dimensionality problem for modelling purposes, so if there are any interaction effects between the features that have not been modelled, due to lack of expertise in this field, there may be considerable amount of underperformance in the model when it is implemented.
- The project assumes that the dataset used for modelling in this project is a representative of the population dataset else the models may not provide the accuracies that are shown here.